Extracellular Vesicles from Human Induced Pluripotent Stem Cells Exhibit a Unique MicroRNA and CircRNA Signature
- PMID: 39664576
- PMCID: PMC11628337
- DOI: 10.7150/ijbs.100113
Extracellular Vesicles from Human Induced Pluripotent Stem Cells Exhibit a Unique MicroRNA and CircRNA Signature
Abstract
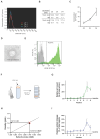
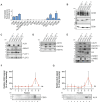
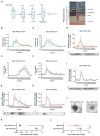
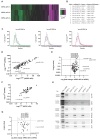
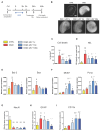
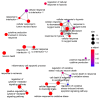
Extracellular vesicles (EV) have emerged as promising cell-free therapeutics in regenerative medicine. However, translating primary cell line-derived EV to clinical applications requires large-scale manufacturing and several challenges, such as replicative senescence, donor heterogeneity, and genetic instability. To address these limitations, we used a reprogramming approach to generate human induced pluripotent stem cells (hiPSC) from the young source of cord blood mesenchymal stem/stromal cells (CBMSC). Capitalizing on their inexhaustible supply potential, hiPSC offer an attractive EV reservoir. Our approach encompassed an exhaustive characterization of hiPSC-EV, aligning with the rigorous MISEV2023 guidelines. Analyses demonstrated physical features compatible with small EV (sEV) and established their identity and purity. Moreover, the sEV-shuttled non-coding (nc) RNA landscape, focusing on the microRNA and circular RNA cargo, completed the molecular signature. The kinetics of the hiPSC-sEV release and cell internalization assays unveiled robust EV production and consistent uptake by human neurons. Furthermore, hiPSC-sEV demonstrated ex vivo cell tissue-protective properties. Finally, via bioinformatics, the potential involvement of the ncRNA cargo in the hiPSC-sEV biological effects was explored. This study significantly advances the understanding of pluripotent stem cell-derived EV. We propose cord blood MSC-derived hiPSC as a promising source for potentially therapeutic sEV.
Keywords: circRNA; cord blood; exosomes; extracellular vesicles; human-induced pluripotent stem cells; miRNA; nanoparticles.
© The author(s).
Conflict of interest statement
Competing Interests: The method for generating human-induced pluripotent stem cells from long-lived cord blood multipotent mesenchymal stromal cells and their use as a source of extracellular vesicles is part of a technology developed by Prof. Lorenza Lazzari and Dr. Mario Barilani. The patent has been granted in Italy (number 102017000141245 on 04/03/2020). The applications for patents in Europe (number EP3720454), US (number US20210161967), and Australia (number AU2018378427) have been filed.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

