Probiotic-facilitated cytokine-induced killer cells suppress peritoneal carcinomatosis and liver metastasis in colorectal cancer cells
- PMID: 39664585
- PMCID: PMC11628340
- DOI: 10.7150/ijbs.101051
Probiotic-facilitated cytokine-induced killer cells suppress peritoneal carcinomatosis and liver metastasis in colorectal cancer cells
Abstract
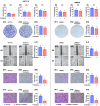
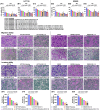
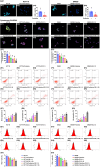
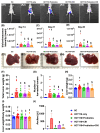
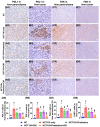
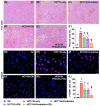
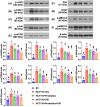
Background: This study tested the hypothesis that combined therapy with probiotics and cytokine-induced killer (CIK) cells was superior to merely one on suppressing the peritoneal carcinomatosis and liver metastasis of colorectal cancer (CRC) cells in nude mice. Methods and Results: The in vitro study revealed that in HCT 116/SW620 CRC cell lines, cell viability, proliferation, colony formation, migratory ability, wound healing, and protein expression of PD-L1 and FAK were significantly and comparably suppressed and that apoptosis was significantly and comparably increased by probiotics and CIK cells, and these effects were further significantly enhanced by combined probiotics + CIK cell therapy (all p<0.001). Nude mice were categorized into Groups 1 (SC), 2 (HCT 116), 3 (HCT 116 + probiotics), 4 (HCT 116 + CIK cells), and 5 (HCT 116 + probiotics + CIK cells). CRC cells were intraperitoneally implanted into Groups 2 to 5, and the animals were euthanized by Day 28. The results demonstrated that the abdominal dissemination of CRC cells, tumor numbers, tumor weights, liver weights, liver necrosis areas and the expression of γ-H2AX/PD-L1/FAK in harvested liver tumors were lowest in Group 1, highest in Group 2, and significantly and progressively decreased in Groups 3 to 5 (all p<0.0001). The protein expression levels of apoptotic and DNA damage biomarkers (Bax/c-caspase 3/c-PARP/γ-H2AX), a metastatic biomarker (FAK) and three tumor proliferation and survival signaling biomarkers (JAK-STAT1, PI3K/Akt/m-TOR and Ras/Raf/MEK/ERK) exhibited identical patterns to that of a tumor immune escape biomarker (PD-L1) among the groups (all p<0.0001). Conclusion: The combination of probiotics and CIK cells was superior to either therapy alone in suppressing CRC cell growth, proliferation, liver metastasis and survival, mainly through downregulating cell proliferation and survival signaling pathways.
Keywords: cell survival signaling; colorectal cancer; cytokine-induced killer cell; liver metastasis; probiotics; tumor mass.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. - PubMed
-
- Sundling KE, Zhang R, Matkowskyj KA. Pathologic Features of Primary Colon, Rectal, and Anal Malignancies. Cancer Treat Res. 2016;168:309–30. - PubMed
-
- Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. - PubMed
-
- Hirasaki Y, Fukunaga M, Sugano M, Nagakari K, Yoshikawa S, Ouchi M. Short- and long-term results of laparoscopic surgery for transverse colon cancer. Surg Today. 2014;44:1266–72. - PubMed
-
- Siani LM, Pulica C. Stage I-IIIC right colonic cancer treated with complete mesocolic excision and central vascular ligation: quality of surgical specimen and long term oncologic outcome according to the plane of surgery. Minerva Chir. 2014;69:199–208. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

