Highlighting chromosomal rearrangements of five species of Galliformes (Domestic fowl, Common and Japanese quail, Barbary and Chukar partridge) and the Houbara bustard, an endangered Otidiformes: banding cytogenetic is a powerful tool
- PMID: 39664601
- PMCID: PMC11632352
- DOI: 10.3897/compcytogen.18.135056
Highlighting chromosomal rearrangements of five species of Galliformes (Domestic fowl, Common and Japanese quail, Barbary and Chukar partridge) and the Houbara bustard, an endangered Otidiformes: banding cytogenetic is a powerful tool
Abstract
Birds are one of the most diverse groups among terrestrial vertebrates. They evolved from theropod dinosaurs, are closely related to the sauropsid group and separated from crocodiles about 240 million years ago. According to the IUCN, 12% of bird populations are threatened with potential extinction. Classical cytogenetics remains a powerful tool for comparing bird genomes and plays a crucial role in the preservation populations of endangered species. It thus makes it possible to detect chromosomal abnormalities responsible for early embryonic mortalities. Thus, in this work, we have provided new information on part of the evolutionary history by analysing high-resolution GTG-banded chromosomes to detect inter- and intrachromosomal rearrangements in six species. Indeed, the first eight autosomal pairs and the sex chromosomes of the domestic fowl Gallusgallusdomesticus Linnaeus, 1758 were compared with five species, four of which represent the order Galliformes (Common and Japanese quail, Gambras and Chukar partridge) and one Otidiformes species (Houbara bustard). Our findings suggest a high degree of conservation of the analysed ancestral chromosomes of the four Galliformes species, with the exception of (double, terminal, para and pericentric) inversions, deletion and the formation of neocentromeres (1, 2, 4, 7, 8, Z and W chromosomes). In addition to the detected rearrangements, reorganisation of the Houbara bustard chromosomes mainly included fusions and fissions involving both macro- and microchromosomes (especially on 2, 4 and Z chromosomes). We also found interchromosomal rearrangements involving shared microchromosomes (10, 11, 13, 14 and 19) between the two analysed avian orders. These rearrangements confirm that the structure of avian karyotypes will be more conserved at the interchromosomal but not at intrachromosomal scale. The appearance ofa small number of inter- and intrachromosomal rearrangements that occurred during evolution suggests a high degree of conservatism of genome organisation in these six species studied. A summary diagram of the rearrangements detected in this study is proposed to explain the chronology of the appearance of various evolutionary events starting from the ancestral karyotype.
Keywords: Avian cytogenetics; GTG-banding; Galliformes; Otidiformes; chromosomal reshuffling; evolution.
Yasmine Kartout-Benmessaoud, Siham Ouchia-Benissad, Leila Mahiddine-Aoudjit, Kafia Ladjali-Mohammedi.
Figures

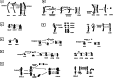

References
-
- Alves Barcellos S, Kretschmer R, Santos de Souza M, Tura V, Pozzobon LC, Ochotorena de Freitas TR, Griffin DK, O’Connor R, Gunski RJ, Del Valle Garnero A. (2024) Understanding microchromosomal organization and evolution in four representative woodpeckers (Picidae, Piciformes) through BAC-FISH analysis. Genome 67(7): 223–232. 10.1139/gen-2023-0096 - DOI - PubMed
-
- Araya-Jaime CA, Silva DMZA, da Silva LRR, do Nascimento CN, Oliveira C, Foresti F. (2022) Karyotype description and comparative chromosomal mapping of rDNA and U2 snDNA sequences in Eigenmannialimbata and E.microstoma (Teleostei, Gymnotiformes, Sternopygidae). Comparative Cytogenetics 16(2): 127–142. 10.3897/compcytogen.v16.i2.72190 - DOI - PMC - PubMed
-
- Azafzaf H, Sande E, Evans SW, Smart M, Collar NJ. (2005) International Action plan for North African Houbara Bustard. A Birdlife International Africa Partnership Publication, 31 pp.
-
- Barcellos SA, Kretschmer R, de Souza MS, Costa AL, Degrandi TM, dos Santos MS, de Oliveira EHC, Cioffi MB, Gunski RJ, Garnero ADV. (2019) Karyotype Evolution and Distinct Evolutionary History of the W Chromosomes in Swallows (Aves, Passeriformes). Cytogenetics and Genome Research 158: 98–105. 10.1159/000500621 - DOI - PubMed
LinkOut - more resources
Full Text Sources
