Finding the last bits of positional information
- PMID: 39664616
- PMCID: PMC11633028
- DOI: 10.1103/prxlife.2.013016
Finding the last bits of positional information
Abstract
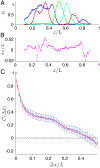
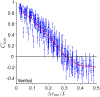
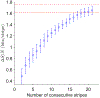
In a developing embryo, information about the position of cells is encoded in the concentrations of morphogen molecules. In the fruit fly, the local concentrations of just a handful of proteins encoded by the gap genes are sufficient to specify position with a precision comparable to the spacing between cells along the anterior-posterior axis. This matches the precision of downstream events such as the striped patterns of expression in the pair-rule genes, but is not quite sufficient to define unique identities for individual cells. We demonstrate theoretically that this information gap can be bridged if positional errors are spatially correlated, with correlation lengths ~ 20% of the embryo length. We then show experimentally that these correlations are present, with the required strength, in the fluctuating positions of the pair-rule stripes, and this can be traced back to the gap genes. Taking account of these correlations, the available information matches the information needed for unique cellular specification, within error bars of ~ 2%. These observation support a precisionist view of information flow through the underlying genetic networks, in which accurate signals are available from the start and preserved as they are transformed into the final spatial patterns.
Figures








Similar articles
-
Scale invariance in early embryonic development.ArXiv [Preprint]. 2023 Dec 29:arXiv:2312.17684v1. ArXiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2403265121. doi: 10.1073/pnas.2403265121. PMID: 38235065 Free PMC article. Updated. Preprint.
-
Positional information, in bits.Proc Natl Acad Sci U S A. 2013 Oct 8;110(41):16301-8. doi: 10.1073/pnas.1315642110. Epub 2013 Oct 2. Proc Natl Acad Sci U S A. 2013. PMID: 24089448 Free PMC article.
-
Modeling the Drosophila pair-rule pattern by reaction-diffusion: gap input and pattern control in a 4-morphogen system.J Theor Biol. 1990 May 22;144(2):171-94. doi: 10.1016/s0022-5193(05)80317-0. J Theor Biol. 1990. PMID: 2197508 Review.
-
Groucho-dependent repression by sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo.Dev Biol. 2004 Dec 15;276(2):541-51. doi: 10.1016/j.ydbio.2004.09.025. Dev Biol. 2004. PMID: 15581884
-
The many bits of positional information.Development. 2021 Feb 1;148(2):dev176065. doi: 10.1242/dev.176065. Development. 2021. PMID: 33526425 Free PMC article. Review.
Cited by
-
Scale invariance in early embryonic development.ArXiv [Preprint]. 2023 Dec 29:arXiv:2312.17684v1. ArXiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2403265121. doi: 10.1073/pnas.2403265121. PMID: 38235065 Free PMC article. Updated. Preprint.
-
Optimization and variability can coexist.ArXiv [Preprint]. 2025 May 29:arXiv:2505.23398v1. ArXiv. 2025. PMID: 40492252 Free PMC article. Preprint.
-
Quantifying the genetic origins of body plan scaling.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2422340121. doi: 10.1073/pnas.2422340121. Epub 2024 Dec 31. Proc Natl Acad Sci U S A. 2025. PMID: 39739820 Free PMC article. No abstract available.
-
Stochastic morphological swings in Hydra regeneration: A manifestation of noisy canalized morphogenesis.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2415736121. doi: 10.1073/pnas.2415736121. Epub 2024 Dec 31. Proc Natl Acad Sci U S A. 2025. PMID: 39739793 Free PMC article.
References
-
- Wolpert L, Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25, 1–47 (1969). - PubMed
-
- Nusslein-Vollhard C and Wieschaus E, Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). - PubMed
-
- Lawrence PA. The Making of a Fly: The Genetics of Animal Design (Blackwell Scientific, Oxford, 1992).
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
