New developments for the Quest for Orthologs benchmark service
- PMID: 39664814
- PMCID: PMC11632614
- DOI: 10.1093/nargab/lqae167
New developments for the Quest for Orthologs benchmark service
Abstract
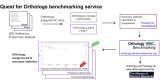
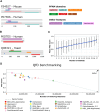
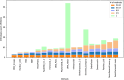
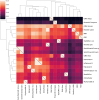
The Quest for Orthologs (QfO) orthology benchmark service (https://orthology.benchmarkservice.org) hosts a wide range of standardized benchmarks for orthology inference evaluation. It is supported and maintained by the QfO consortium, and is used to gather ortholog predictions and to examine strengths and weaknesses of newly developed and existing orthology inference methods. The web server allows different inference methods to be compared in a standardized way using the same proteome data. The benchmark results are useful for developing new methods and can help researchers to guide their choice of orthology method for applications in comparative genomics and phylogenetic analysis. We here present a new release of the Orthology Benchmark Service with a new benchmark based on feature architecture similarity as well as updated reference proteomes. We further provide a meta-analysis of the public predictions from 18 different orthology assignment methods to reveal how they relate in terms of ortholog predictions and benchmark performance. These results can guide users of orthologs to the best suited method for their purpose.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures
References
-
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970; 19:99–113. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

