GIN-TONIC: non-hierarchical full-text indexing for graph genomes
- PMID: 39664816
- PMCID: PMC11632618
- DOI: 10.1093/nargab/lqae159
GIN-TONIC: non-hierarchical full-text indexing for graph genomes
Abstract
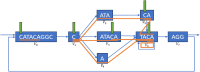
This paper presents a new data structure, GIN-TONIC (Graph INdexing Through Optimal Near Interval Compaction), designed to index arbitrary string-labelled directed graphs representing, for instance, pangenomes or transcriptomes. GIN-TONIC provides several capabilities not offered by other graph-indexing methods based on the FM-Index. It is non-hierarchical, handling a graph as a monolithic object; it indexes at nucleotide resolution all possible walks in the graph without the need to explicitly store them; it supports exact substring queries in polynomial time and space for all possible walk roots in the graph, even if there are exponentially many walks corresponding to such roots. Specific ad-hoc optimizations, such as precomputed caches, allow GIN-TONIC to achieve excellent performance for input graphs of various topologies and sizes. Robust scalability capabilities and a querying performance close to that of a linear FM-Index are demonstrated for two real-world applications on the scale of human pangenomes and transcriptomes. Source code and associated benchmarks are available on GitHub.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


References
LinkOut - more resources
Full Text Sources

