Platelet-Monocyte Aggregate Instigates Inflammation and Vasculopathy in Kawasaki Disease
- PMID: 39665236
- PMCID: PMC11792051
- DOI: 10.1002/advs.202406282
Platelet-Monocyte Aggregate Instigates Inflammation and Vasculopathy in Kawasaki Disease
Abstract
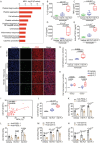
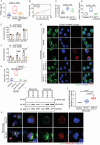
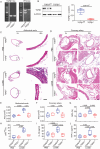
Kawasaki disease (KD) is a severe acute febrile illness and systemic vasculitis that causes coronary artery aneurysms in young children. Platelet hyperreactivity and an aberrant immune response are key indicators of KD; however, the mechanism by which hyperactive platelets contribute to inflammation and vasculopathy in KD remains unclear. A cytokine-mediated positive feedback loop between KD platelets and monocytes is identified. KD platelet-monocyte aggregates (MPAs) are mediated by an initial interaction of P-selectin (cluster of differentiation 62P, CD62p) and its glycoprotein ligand 1 (PSGL-1). This is followed by a coordinated interaction of platelet glycoprotein (GP)Ibα with monocyte CD11b. Monocyte-activated platelets initiate transforming growth factor (TGF)β1 release, which results in nuclear localization of nuclear factor kappaB in monocytes, therefore, driving the phenotypic conversion of classical monocytes (CD14+CD16-) into proinflammatory monocytes (CD14+CD16+). The platelet-activated monocytes release interleukin-1 and tissue necrotic factor-α, which promote further platelet activation. KD-induced inflammation and vasculopathy are prevented by inhibiting the components of this positive feedback loop. Notably, mice deficient in platelet TGFβ1 show less MPA and CD14+CD16+ monocytes, along with reduced inflammation and vasculopathy. These findings reveal that platelet-monocyte interactive proteins (CD62p/PSGL-1 and (GP)Ibα/CD11b) and cytokine mediators (platelet TGFβ1) are potential biomarkers and therapeutic targets for KD vasculopathy.
Keywords: Kawasaki disease; inflammation; monocyte; platelet; vasculopathy.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
- 2024A1515030045/National Science Foundation of Guangdong province
- 202201020651/Guangzhou Basic and applied basic research
- 2024A04J6579/Guangzhou Basic and applied basic research
- 2024A03J1089/Guangzhou School (Institute) Enterprise Joint Funding Project-Outstanding talent project
- CP/2023/2.1/Research Matching Grant of Hong Kong Metropolitan University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
