Arabidopsis WALL-ASSOCIATED KINASES are not required for oligogalacturonide-induced signaling and immunity
- PMID: 39665686
- PMCID: PMC11684076
- DOI: 10.1093/plcell/koae317
Arabidopsis WALL-ASSOCIATED KINASES are not required for oligogalacturonide-induced signaling and immunity
Abstract
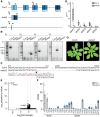
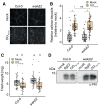
Carbohydrate-based cell wall signaling impacts plant growth, development, and stress responses; however, how cell wall signals are perceived and transduced remains poorly understood. Several cell wall breakdown products have been described as typical damage-associated molecular patterns that activate plant immunity, including pectin-derived oligogalacturonides (OGs). Receptor kinases of the WALL-ASSOCIATED KINASE (WAK) family bind pectin and OGs and were previously proposed as OG receptors. However, unambiguous genetic evidence for the role of WAKs in OG responses is lacking. Here, we investigated the role of Arabidopsis (Arabidopsis thaliana) WAKs in OG perception using a clustered regularly interspaced short palindromic repeats mutant in which all 5 WAK genes were deleted. Using a combination of immune assays for early and late pattern-triggered immunity, we show that WAKs are dispensable for OG-induced signaling and immunity, indicating that they are not bona fide OG receptors.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



References
-
- Bender KW, Couto D, Kadota Y, Macho AP, Sklenar J, Derbyshire P, Bjornson M, DeFalco TA, Petriello A, Font Farre M, et al. . Activation loop phosphorylation of a non-RD receptor kinase initiates plant innate immune signaling. Proc Natl Acad Sci U S A. 2021:118(38):e2108242118. 10.1073/pnas.2108242118 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

