Alpha-1 Antitrypsin Inclusions Sequester GRP78 in a Bile Acid-Inducible Manner
- PMID: 39665869
- PMCID: PMC11636636
- DOI: 10.1111/liv.16207
Alpha-1 Antitrypsin Inclusions Sequester GRP78 in a Bile Acid-Inducible Manner
Abstract
Background and aims: The homozygous PiZ mutation (PIZZ genotype) constitutes the predominant cause of severe alpha-1 antitrypsin (AAT) deficiency and leads to liver disease via hepatocellular AAT aggregation. We systematically analysed the composition of AAT aggregates and studied the impact of bile acids.
Methods: AAT inclusions were isolated from livers of PiZ overexpressing mice and PIZZ humans via fluorescence-activated and immunomagnetic sorting (FACS/MACS), while insoluble proteins were obtained via Triton-X extraction. Inclusion composition was evaluated through mass-spectrometry (MS), immunoblotting and immunostaining. Hepatocytes with versus without AAT aggregates were obtained via microdissection. Serum bile acids were assessed in 57 PIZZ subjects and 19 controls. Mice were administered 2% cholic acid (CA)-supplemented chow for 7 days.
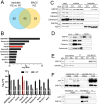
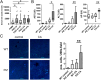
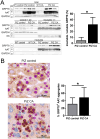
Results: MS identified the key endoplasmic reticulum chaperone 78 kDa glucose-regulated protein (GRP78) in FACS/MACS pulldowns. GRP78 was also enriched in insoluble fractions from PiZ mice versus wild types and detected in insoluble fractions/MACS isolates from PIZZ liver explants. In cultured cells/primary hepatocytes, PiZ overexpression was associated with increased GRP78 mRNA/protein levels. In human livers, hepatocytes with AAT aggregates had higher GRP78 levels than hepatocytes without. PIZZ subjects displayed higher serum bile acid levels than controls and the highest levels were seen in individuals with liver injury/fibrosis. In PiZ mice, CA-mediated bile acid challenge resulted in increased liver injury and translocation of GRP78 into the aggregates.
Conclusions: Our results demonstrate that GRP78 is sequestered within AAT inclusions. Bile acid accumulation, as seen in PIZZ subjects with liver disease, may promote GRP78 segregation and thereby augment liver damage.
Trial registration: NCT02929940.
Keywords: ER chaperone; cholestatic liver injury; genetic liver disease; rare liver disease; α‐1 antitrypsin deficiency.
© 2024 The Author(s). Liver International published by John Wiley & Sons Ltd.
Conflict of interest statement
P.S. reports receiving grants and honoraria from Arrowhead Pharmaceuticals, CSL Behring, Grifols Inc., consulting fees or honoraria from Alnylam Pharmaceuticals, Arrowhead Pharmaceuticals, BioMarin Pharmaceutical, BridgeBio, Dicerna Pharmaceuticals, GSK, Intellia Pharmaceuticals, Takeda Pharmaceuticals, Novo Nordisk and Ono Pharmaceuticals, participating in leadership or fiduciary roles in Alpha1‐Deutschland, Alpha1 Global and material transfer support for Vertex Pharmaceuticals and Dicerna Pharmaceuticals.
Figures


References
-
- Chiti F. and Dobson C. M., “Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade,” Annual Review of Biochemistry 86 (2017): 27–68. - PubMed
-
- Lee W.‐C. M., Yoshihara M., and Littleton J. T., “Cytoplasmic Aggregates Trap Polyglutamine‐Containing Proteins and Block Axonal Transport in a Drosophila Model of Huntington's Disease,” Proceedings of the National Academy of Sciences of the United States of America 101 (2004): 3224–3229. - PMC - PubMed
-
- Strnad P., McElvaney N. G., and Lomas D. A., “Alpha1‐Antitrypsin Deficiency,” New England Journal of Medicine 382 (2020): 1443–1455. - PubMed
-
- Lomas D. A., Evans D. L., Finch J. T., and Carrell R. W., “The Mechanism of Z Alpha 1‐Antitrypsin Accumulation in the Liver,” Nature 357 (1992): 605–607. - PubMed
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

