Red blood cells capture and deliver bacterial DNA to drive host responses during polymicrobial sepsis
- PMID: 39666381
- PMCID: PMC11827885
- DOI: 10.1172/JCI182127
Red blood cells capture and deliver bacterial DNA to drive host responses during polymicrobial sepsis
Abstract
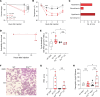
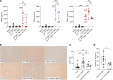
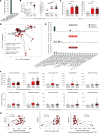
Red blood cells (RBCs), traditionally recognized for their role in transporting oxygen, play a pivotal role in the body's immune response by expressing TLR9 and scavenging excess host cell-free DNA. DNA capture by RBCs leads to accelerated RBC clearance and triggers inflammation. Whether RBCs can also acquire microbial DNA during infections is unknown. Murine RBCs acquire microbial DNA in vitro, and bacterial DNA-induced (bDNA-induced) macrophage activation was augmented by WT, but not Tlr9-deleted, RBCs. In a mouse model of polymicrobial sepsis, RBC-bound bDNA was elevated in WT mice but not in erythroid Tlr9-deleted mice. Plasma cytokine analysis in these mice revealed distinct sepsis clusters characterized by persistent hypothermia and hyperinflammation in the most severely affected mice. RBC Tlr9 deletion attenuated plasma and tissue IL-6 production in the most severely affected group. Parallel findings in humans confirmed that RBCs from patients with sepsis harbored more bDNA than did RBCs from healthy individuals. Further analysis through 16S sequencing of RBC-bound DNA illustrated distinct microbial communities, with RBC-bound DNA composition correlating with plasma IL-6 in patients with sepsis. Collectively, these findings unveil RBCs as overlooked reservoirs and couriers of microbial DNA, capable of influencing host inflammatory responses in sepsis.
Keywords: Cytokines; Inflammation; Innate immunity; Pulmonology.
Conflict of interest statement
Figures

Update of
-
Red Blood Cell DNA Capture and Delivery Drives Host Responses During Polymicrobial Sepsis.Res Sq [Preprint]. 2024 Apr 23:rs.3.rs-2592196. doi: 10.21203/rs.3.rs-2592196/v2. Res Sq. 2024. Update in: J Clin Invest. 2024 Dec 12;135(4):e182127. doi: 10.1172/JCI182127. PMID: 38746470 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

