Utilizing AI for the Identification and Validation of Novel Therapeutic Targets and Repurposed Drugs for Endometriosis
- PMID: 39666559
- PMCID: PMC11792045
- DOI: 10.1002/advs.202406565
Utilizing AI for the Identification and Validation of Novel Therapeutic Targets and Repurposed Drugs for Endometriosis
Abstract
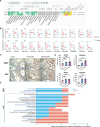
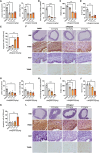
Endometriosis affects over 190 million women globally, and effective therapies are urgently needed to address the burden of endometriosis on women's health. Using an artificial intelligence (AI)-driven target discovery platform, two unreported therapeutic targets, guanylate-binding protein 2 (GBP2) and hematopoietic cell kinase (HCK) are identified, along with a drug repurposing target, integrin beta 2 (ITGB2) for the treatment of endometriosis. GBP2, HCK, and ITGB2 are upregulated in human endometriotic specimens. siRNA-mediated knockdown of GBP2 and HCK significantly reduced cell viability and proliferation while stimulating apoptosis in endometrial stromal cells. In subcutaneous and intraperitoneal endometriosis mouse models, siRNAs targeting GBP2 and HCK notably reduced lesion volume and weight, with decreased proliferation and increased apoptosis within lesions. Both subcutaneous and intraperitoneal administration of Lifitegrast, an approved ITGB2 antagonist, effectively suppresses lesion growth. Collectively, these data present Lifitegrast as a previously unappreciated intervention for endometriosis treatment and identify GBP2 and HCK as novel druggable targets in endometriosis treatment. This study underscores AI's potential to accelerate the discovery of novel drug targets and facilitate the repurposing of treatment modalities for endometriosis.
Keywords: GBP2; HCK; ITGB2; artificial intelligence; drug repurposing; endometriosis; lifitegrast; target discovery.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
B.H.M.L., X.L., A.G., F.R., A.Z., and F.W.P. are affiliated with Insilico Medicine, a commercial company developing AI solutions for aging research, drug discovery, and longevity medicine. Y.L., S.W.H., and C.C.W. declare no competing interests.
Figures




References
-
- Moingeon P., Ann. Pharm. Fr. 2021, 79, 566. - PubMed
-
- Swinney D. C., Anthony J., Nat. Rev. Drug Discovery 2011, 10, 507. - PubMed
-
- Plenge R. M., Scolnick E. M., Altshuler D., Nat. Rev. Drug Discovery 2013, 12, 581. - PubMed
-
- Zhavoronkov A., Ivanenkov Y. A., Aliper A., Veselov M. S., Aladinskiy V. A., Aladinskaya A. V., Terentiev V. A., Polykovskiy D. A., Kuznetsov M. D., Asadulaev A., Volkov Y., Zholus A., Shayakhmetov R. R., Zhebrak A., Minaeva L. I., Zagribelnyy B. A., Lee L. H., Soll R., Madge D., Xing L., Guo T., Aspuru‐Guzik A., Nat. Biotechnol. 2019, 37, 1038. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
