Seasonal influenza a virus lineages exhibit divergent abilities to antagonize interferon induction and signaling
- PMID: 39666644
- PMCID: PMC11637315
- DOI: 10.1371/journal.ppat.1012727
Seasonal influenza a virus lineages exhibit divergent abilities to antagonize interferon induction and signaling
Abstract
The circulation of seasonal influenza A viruses (IAVs) in humans relies on effective evasion and subversion of the host immune response. While the evolution of seasonal H1N1 and H3N2 viruses to avoid humoral immunity is well characterized, relatively little is known about the evolution of innate immune antagonism phenotypes in these viruses. Numerous studies have established that only a small subset of infected cells is responsible for initiating the type I and type III interferon (IFN) response during IAV infection, emphasizing the importance of single cell studies to accurately characterize the IFN response during infection. We developed a flow cytometry-based method to examine transcriptional changes in IFN and interferon stimulated gene (ISG) expression at the single cell level. We observed that NS segments derived from seasonal H3N2 viruses are more efficient at antagonizing IFN signaling but less effective at suppressing IFN induction, compared to the pdm2009 H1N1 lineage. We compared a collection of NS segments spanning the natural history of the current seasonal IAV lineages and demonstrate long periods of stability in IFN antagonism potential, punctuated by occasional phenotypic shifts. Altogether, our data reveal significant differences in how seasonal and pandemic H1N1 and H3N2 viruses antagonize the human IFN response at the single cell level.
Copyright: © 2024 Rivera-Cardona et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
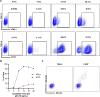
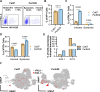




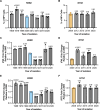
Update of
-
Seasonal influenza A virus lineages exhibit divergent abilities to antagonize interferon induction and signaling.bioRxiv [Preprint]. 2024 Jul 12:2024.07.11.603146. doi: 10.1101/2024.07.11.603146. bioRxiv. 2024. Update in: PLoS Pathog. 2024 Dec 12;20(12):e1012727. doi: 10.1371/journal.ppat.1012727. PMID: 39026731 Free PMC article. Updated. Preprint.
References
-
- Stifter SA, Bhattacharyya N, Pillay R, Flórido M, Triccas JA, Britton WJ, et al.. Functional Interplay between Type I and II Interferons Is Essential to Limit Influenza A Virus-Induced Tissue Inflammation. PLOS Pathogens. 2016. Jan 5;12(1):e1005378. doi: 10.1371/journal.ppat.1005378 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

