Novel kinase regulators of extracellular matrix internalisation identified by high-content screening modulate invasive carcinoma cell migration
- PMID: 39666682
- PMCID: PMC11637276
- DOI: 10.1371/journal.pbio.3002930
Novel kinase regulators of extracellular matrix internalisation identified by high-content screening modulate invasive carcinoma cell migration
Abstract
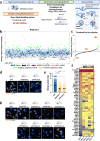
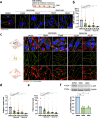
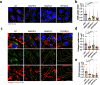
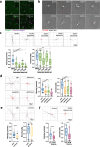
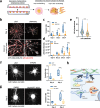
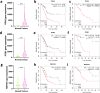
The interaction between cancer cells and the extracellular matrix (ECM) plays a pivotal role in tumour progression. While the extracellular degradation of ECM proteins has been well characterised, ECM endocytosis and its impact on cancer cell progression, migration, and metastasis is poorly understood. ECM internalisation is increased in invasive breast cancer cells, suggesting it may support invasiveness. However, current high-throughput approaches mainly focus on cells grown on plastic in 2D, making it difficult to apply these to the study of ECM dynamics. Here, we developed a high-content screening assay to study ECM uptake, based on the of use automated ECM coating for the generation of highly homogeneous ECM a pH-sensitive dye to image ECM trafficking in live cells. We identified that mitogen-activated protein kinase (MAPK) family members, MAP3K1 and MAPK11 (p38β), and the protein phosphatase 2 (PP2) subunit PPP2R1A were required for the internalisation of ECM-bound α2β1 integrin. Mechanistically, we show that down-regulation of the sodium/proton exchanger 1 (NHE1), an established macropinocytosis regulator and a target of p38, mediated ECM macropinocytosis. Moreover, disruption of α2 integrin, MAP3K1, MAPK11, PPP2R1A, and NHE1-mediated ECM internalisation significantly impaired cancer cell migration and invasion in 2D and 3D culture systems. Of note, integrin-bound ECM was targeted for lysosomal degradation, which was required for cell migration on cell-derived matrices. Finally, α2β1 integrin and MAP3K1 expression were significantly up-regulated in pancreatic tumours and correlated with poor prognosis in pancreatic cancer patients. Strikingly, MAP3K1, MAPK11, PPP2R1A, and α2 integrin expression were higher in chemotherapy-resistant tumours in breast cancer patients. Our results identified the α2β1 integrin/p38 signalling axis as a novel regulator of ECM endocytosis, which drives invasive migration and tumour progression, demonstrating that our high-content screening approach has the capability of identifying novel regulators of cancer cell invasion.
Copyright: © 2024 Martinez et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

