Diverse phage communities are maintained stably on a clonal bacterial host
- PMID: 39666794
- PMCID: PMC7617280
- DOI: 10.1126/science.adk1183
Diverse phage communities are maintained stably on a clonal bacterial host
Abstract
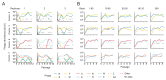
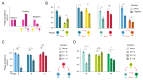
Bacteriophages are the most abundant and phylogenetically diverse biological entities on Earth, yet the ecological mechanisms that sustain this extraordinary diversity remain unclear. In this study, we discovered that phage diversity consistently outstripped the diversity of their bacterial hosts under simple experimental conditions. We assembled and passaged dozens of diverse phage communities on a single, nonevolving strain of Escherichia coli until the phage communities reached equilibrium. In all cases, we found that two or more phage species coexisted stably, despite competition for a single, clonal host population. Phage coexistence was supported through host phenotypic heterogeneity, whereby bacterial cells adopting different growth phenotypes served as niches for different phage species. Our experiments reveal that a rich community ecology of bacteriophages can emerge on a single bacterial host.
Conflict of interest statement
Figures


References
-
- Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, McDonnell SA, Khokhlova EV, Draper LA, Forde A, Guerin E, et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe. 2019;26:527–541.:e5. - PubMed
-
- Neri U, Wolf YI, Roux S, Camargo AP, Lee B, Kazlauskas D, Chen IM, Ivanova N, Zeigler Allen L, Paez-Espino D, Bryant DA, et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell. 2022;185:4023–4037.:e18. - PubMed
-
- Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory. Annu Rev Virol. 2017;4:201–219. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

