A near-tight lower bound on the density of forward sampling schemes
- PMID: 39666942
- PMCID: PMC11676336
- DOI: 10.1093/bioinformatics/btae736
A near-tight lower bound on the density of forward sampling schemes
Abstract
Motivation: Sampling k-mers is a ubiquitous task in sequence analysis algorithms. Sampling schemes such as the often-used random minimizer scheme are particularly appealing as they guarantee at least one k-mer is selected out of every w consecutive k-mers. Sampling fewer k-mers often leads to an increase in efficiency of downstream methods. Thus, developing schemes that have low density, i.e. have a small proportion of sampled k-mers, is an active area of research. After over a decade of consistent efforts in both decreasing the density of practical schemes and increasing the lower bound on the best possible density, there is still a large gap between the two.
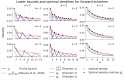
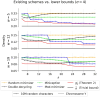
Results: We prove a near-tight lower bound on the density of forward sampling schemes, a class of schemes that generalizes minimizer schemes. For small w and k, we observe that our bound is tight when k≡1(mod w). For large w and k, the bound can be approximated by 1w+k⌈w+kw⌉. Importantly, our lower bound implies that existing schemes are much closer to achieving optimal density than previously known. For example, with the current default minimap2 HiFi settings w = 19 and k = 19, we show that the best known scheme for these parameters, the double decycling-set-based minimizer of Pellow et al. is at most 3% denser than optimal, compared to the previous gap of at most 50%. Furthermore, when k≡1(mod w) and the alphabet size σ goes to ∞, we show that mod-minimizers introduced by Groot Koerkamp and Pibiri achieve optimal density matching our lower bound.
Availability and implementation: Minimizer implementations: github.com/RagnarGrootKoerkamp/minimizers ILP and analysis: github.com/treangenlab/sampling-scheme-analysis.
© The Author(s) 2024. Published by Oxford University Press.
Figures

Update of
-
A near-tight lower bound on the density of forward sampling schemes.bioRxiv [Preprint]. 2024 Nov 19:2024.09.06.611668. doi: 10.1101/2024.09.06.611668. bioRxiv. 2024. Update in: Bioinformatics. 2024 Dec 26;41(1):btae736. doi: 10.1093/bioinformatics/btae736. PMID: 39605515 Free PMC article. Updated. Preprint.
References
-
- DeBlasio D, Gbosibo F, Kingsford C. et al. Practical universal k-mer sets for minimizer schemes. In: Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Niagara Falls, NY, USA. New York, NY, USA: Association for Computing Machinery, 2019, 167–76. 10.1145/3307339.3342144 - DOI
-
- Ekim B, Berger B, Orenstein Y.. A randomized parallel algorithm for efficiently finding near-optimal universal hitting sets. In: International Conference on Research in Computational Molecular Biology, Padua, Italy. Cham, Switzerland: Springer International Publishing, 2020, 37–53. 10.1007/978-3-030-45257-5_3 - PMC - PubMed
-
- Golan S, Tziony I, Kraus M. et al. Generating low-density minimizers. bioRxiv, 2024. 10.1101/2024.10.28.620726, preprint: not peer reviewed. - DOI
-
- Groot Koerkamp R, Pibiri GE. The mod-minimizer: a simple and efficient sampling algorithm for long k-mers. In: Pissis SP, Sung WK (eds), 24th International Workshop on Algorithms in Bioinformatics (WABI 2024), volume 312 of Leibniz International Proceedings in Informatics (LIPIcs). Dagstuhl, Germany: Schloss Dagstuhl—Leibniz-Zentrum für Informatik, 2024, 11:1–11:23. ISBN 978-3-95977-340-9. 10.4230/LIPIcs.WABI.2024.11. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

