Molecular features of the serological IgG repertoire elicited by egg-based, cell-based, or recombinant haemagglutinin-based seasonal influenza vaccines: a comparative, prospective, observational cohort study
- PMID: 39667375
- PMCID: PMC11807745
- DOI: 10.1016/j.lanmic.2024.06.002
Molecular features of the serological IgG repertoire elicited by egg-based, cell-based, or recombinant haemagglutinin-based seasonal influenza vaccines: a comparative, prospective, observational cohort study
Abstract
Background: Egg-based inactivated quadrivalent seasonal influenza vaccine (eIIV4), cell culture-based inactivated quadrivalent seasonal influenza vaccine (ccIIV4), and recombinant haemagglutinin (HA)-based quadrivalent seasonal influenza vaccine (RIV4) have been licensed for use in the USA. In this study, we used antigen-specific serum proteomics analysis to assess how the molecular composition and qualities of the serological antibody repertoires differ after seasonal influenza immunisation by each of the three vaccines and how different vaccination platforms affect the HA binding affinity and breadth of the serum antibodies that comprise the polyclonal response.
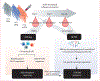
Methods: In this comparative, prospective, observational cohort study, we included female US health-care personnel (mean age 47·6 years [SD 8]) who received a single dose of RIV4, eIIV4, or ccIIV4 during the 2018-19 influenza season at Baylor Scott & White Health (Temple, TX, USA). Eligible individuals were selected based on comparable day 28 serum microneutralisation titres and similar vaccination history. Laboratory investigators were blinded to assignment until testing was completed. The preplanned exploratory endpoints were assessed by deconvoluting the serological repertoire specific to A/Singapore/INFIMH-16-0019/2016 (H3N2) HA before (day 0) and after (day 28) immunisation using bottom-up liquid chromatography-mass spectrometry proteomics (referred to as Ig-Seq) and natively paired variable heavy chain-variable light chain high-throughput B-cell receptor sequencing (referred to as BCR-Seq). Features of the antigen-specific serological repertoire at day 0 and day 28 for the three vaccine groups were compared. Antibodies identified with high confidence in sera were recombinantly expressed and characterised in depth to determine the binding affinity and breadth to time-ordered H3 HA proteins.
Findings: During September and October of the 2018-19 influenza season, 15 individuals were recruited and assigned to receive RIV4 (n=5), eIIV4 (n=5), or ccIIV4 (n=5). For all three cohorts, the serum antibody repertoire was dominated by back-boosted antibody lineages (median 98% [95% CI 88-99]) that were present in the serum before vaccination. Although vaccine platform-dependent differences were not evident in the repertoire diversity, somatic hypermutation, or heavy chain complementarity determining region 3 biochemical features, antibodies boosted by RIV4 showed substantially higher binding affinity to the vaccine H3/HA (median half-maximal effective concentration [EC50] to A/Singapore/INFIMH-16-0019/2016 HA: 0·037 μg/mL [95% CI 0·012-0·12] for RIV4; 4·43 μg/mL [0·030-100·0] for eIIV4; and 18·50 μg/mL [0·99-100·0] μg/mL for ccIIV4) and also the HAs from contemporary H3N2 strains than did those elicited by eIIV4 or ccIIV4 (median EC50 to A/Texas/50/2012 HA: 0·037 μg/mL [0·017-0·32] for RIV4; 1·10 μg/mL [0·045-100] for eIIV4; and 12·6 μg/mL [1·8-100] for ccIIV4). Comparison of B-cell receptor sequencing repertoires on day 7 showed that eIIV4 increased the median frequency of canonical egg glycan-targeting B cells (0·20% [95% CI 0·067-0·37] for eIIV4; 0·058% [0·050-0·11] for RIV4; and 0·035% [0-0·062] for ccIIV4), whereas RIV4 vaccination decreased the median frequency of B-cell receptors displaying stereotypical features associated with membrane proximal anchor-targeting antibodies (0·062% [95% CI 0-0·084] for RIV4; 0·12% [0·066-0·16] for eIIV4; and 0·18% [0·016-0·20] for ccIIV4). In exploratory analysis, we characterised the structure of a highly abundant monoclonal antibody that binds to both group 1 and 2 HAs and recognises the HA trimer interface, despite its sequence resembling the stereotypical sequence motif found in membrane-proximal anchor binding antibodies.
Interpretation: Although all three licensed seasonal influenza vaccines elicit serological antibody repertoires with indistinguishable features shaped by heavy imprinting, the RIV4 vaccine selectively boosts higher affinity monoclonal antibodies to contemporary strains and elicits greater serum binding potency and breadth, possibly as a consequence of the multivalent structural features of the HA immunogen in this vaccine formulation. Collectively, our findings show advantages of RIV4 vaccines and more generally highlight the benefits of multivalent HA immunogens in promoting higher affinity serum antibody responses.
Funding: Centers for Disease Control and Prevention, National Institutes of Health, and Bill & Melinda Gates Foundation.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests GG receives royalties or compensation from Amgen, Asher Bio, Grifols, Cell Signaling Technologies, and Texas A&M University, and reports grant support from Clayton Foundation for Research, none of which poses a conflict of interest with this work. MG reports contracts from CDC (US Flu VE Network, HAIVEN, Synergy study), CDC-Abt Associates (RECOVER-PROJECT Cohort studies), CDC-Vanderbilt (IVY Network), and CDC-Westat (VISION study). MG receives honoraria from CDC–Texas Chapter of the American Academy of Pediatrics (AAP)–Texas Pediatric Society (TPS) Project Firstline. MG is co-chair of the Infectious Diseases and Immunization Committee and is chair of the Texas RSV Task Force from TPS, AAP. All other authors declare no competing interests.
Figures





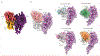
References
-
- Wang W, Alvarado-Facundo E, Vassell R, et al. Comparison of A(H3N2) neutralizing antibody responses elicited by 2018–2019 season quadrivalent influenza vaccines derived from eggs, cells, and recombinant hemagglutinin. Clin Infect Dis 2021; 73: e4312–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

