Inhibition of the PI3K-AKT-MTORC1 axis reduces the burden of the m.3243A>G mtDNA mutation by promoting mitophagy and improving mitochondrial function
- PMID: 39667405
- PMCID: PMC11925111
- DOI: 10.1080/15548627.2024.2437908
Inhibition of the PI3K-AKT-MTORC1 axis reduces the burden of the m.3243A>G mtDNA mutation by promoting mitophagy and improving mitochondrial function
Abstract
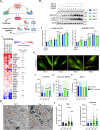
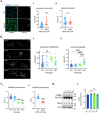
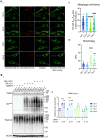
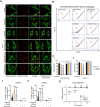
Mitochondrial DNA (mtDNA) encodes genes essential for oxidative phosphorylation. The m.3243A>G mutation causes severe disease, including myopathy, lactic acidosis and stroke-like episodes (MELAS) and is the most common pathogenic mtDNA mutation in humans. We have previously shown that the mutation is associated with constitutive activation of the PI3K-AKT-MTORC1 axis. Inhibition of this pathway in patient fibroblasts reduced the mutant load, rescued mitochondrial bioenergetic function and reduced glucose dependence. We have now investigated the mechanisms that select against the mutant mtDNA under these conditions. Basal macroautophagy/autophagy and lysosomal degradation of mitochondria were suppressed in the mutant cells. Pharmacological inhibition of any step of the PI3K-AKT-MTORC1 pathway activated mitophagy and progressively reduced m.3243A>G mutant load over weeks. Inhibition of autophagy with bafilomycin A1 or chloroquine prevented the reduction in mutant load, suggesting that mitophagy was necessary to remove the mutant mtDNA. Inhibition of the pathway was associated with metabolic remodeling - mitochondrial membrane potential and respiratory rate improved even before a measurable fall in mutant load and proved crucial for mitophagy. Thus, maladaptive activation of the PI3K-AKT-MTORC1 axis and impaired autophagy play a major role in shaping the presentation and progression of disease caused by the m.3243A>G mutation. Our findings highlight a potential therapeutic target for this otherwise intractable disease.Abbreviation: ΔΨm: mitochondrial membrane potential; 2DG: 2-deoxy-D-glucose; ANOVA: analysis of variance; ARMS-qPCR: amplification-refractory mutation system quantitative polymerase chain reaction; Baf A1: bafilomycin A1; BSA: bovine serum albumin; CQ: chloroquine; Cybrid: cytoplasmic hybrid; CYCS: cytochrome c, somatic; DCA: dichloroacetic acid; DMEM: Dulbecco's modified Eagle's medium; DMSO: dimethylsulfoxide; EGFP: enhanced green fluorescent protein; LC3B-I: carboxy terminus cleaved microtubule-associated protein 1 light chain 3 beta; LC3B-II: lipidated microtubule-associated protein 1 light chain 3 beta; LY: LY290042; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MELAS: mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes; MFC: mitochondrial fragmentation count; mt-Keima: mitochondrial-targeted mKeima; mtDNA: mitochondrial DNA/mitochondrial genome; MTOR: mechanistic target of rapamycin kinase; MTORC1: MTOR complex 1; OA: oligomycin+antimycin A; OxPhos: oxidative phosphorylation; DPBS: Dulbecco's phosphate-buffered saline; PPARGC1A/PGC-1α: PPARG coactivator 1 alpha; PPARGC1B/PGC-1β: PPARG coactivator 1 beta; PI3K: phosphoinositide 3-kinase; PINK1: PTEN induced kinase 1; qPCR: quantitative polymerase chain reaction; RNA-seq: RNA sequencing; RP: rapamycin; SQSTM1/p62: sequestosome 1; TEM: transmission electron microscopy; WT: wild-type.
Keywords: PI3K-AKT-MTORC1; m.3243A>G; mitochondria; mitophagy; mtDNA mutations; nutrient signaling.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
