Earliest modern human genomes constrain timing of Neanderthal admixture
- PMID: 39667410
- PMCID: PMC11839475
- DOI: 10.1038/s41586-024-08420-x
Earliest modern human genomes constrain timing of Neanderthal admixture
Abstract
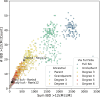
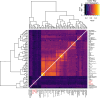
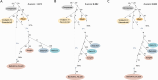
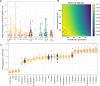
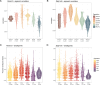
Modern humans arrived in Europe more than 45,000 years ago, overlapping at least 5,000 years with Neanderthals1-4. Limited genomic data from these early modern humans have shown that at least two genetically distinct groups inhabited Europe, represented by Zlatý kůň, Czechia3 and Bacho Kiro, Bulgaria2. Here we deepen our understanding of early modern humans by analysing one high-coverage genome and five low-coverage genomes from approximately 45,000-year-old remains from Ilsenhöhle in Ranis, Germany4, and a further high-coverage genome from Zlatý kůň. We show that distant familial relationships link the Ranis and Zlatý kůň individuals and that they were part of the same small, isolated population that represents the deepest known split from the Out-of-Africa lineage. Ranis genomes harbour Neanderthal segments that originate from a single admixture event shared with all non-Africans that we date to approximately 45,000-49,000 years ago. This implies that ancestors of all non-Africans sequenced so far resided in a common population at this time, and further suggests that modern human remains older than 50,000 years from outside Africa represent different non-African populations.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

