Improved influenza A whole-genome sequencing protocol
- PMID: 39669272
- PMCID: PMC11635996
- DOI: 10.3389/fcimb.2024.1497278
Improved influenza A whole-genome sequencing protocol
Abstract
Influenza A virus poses significant public health challenges due to its high mutation rate and zoonotic potential. Whole-genome sequencing (WGS) is crucial for monitoring and characterizing these viruses. Oxford Nanopore Technologies (ONT) and Illumina next-generation sequencing platforms are commonly used, with ONT being advantageous for its long-read capabilities, portability, and unique ability to access raw data in real-time during sequencing, making it suitable for rapid outbreak responses. This study optimizes the ONT Ligation Sequencing Influenza A Whole Genome protocol by refining RT-PCR kits, primers, and purification methods, and evaluating automation for high-throughput processing. The alternative RT-PCR kits, combined with alternative primers, significantly improved read depth coverage and reduced short, untargeted reads compared to the original ONT protocol. The improvement was particularly evident in the minimum read depth coverage of polymerase segments, which often face challenges with achieving uniform coverage, displaying higher coverage at the 5' and 3' termini, and lower coverage in the central regions. This optimized protocol for targeted influenza A WGS not only enhances sequencing quality and efficiency, but is applicable to all NGS platforms, making it highly valuable for studying influenza adaptation and improving surveillance. Additionally, this protocol can be further refined and adapted for the sequencing of other pathogens, broadening its utility in various pathogen monitoring and response efforts.
Keywords: Illumina; MinION; NGS; RT-PCR; WGS; influenza; nanopore; next-generation sequencing.
Copyright © 2024 Goraichuk, Risalvato, Pantin-Jackwood and Suarez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
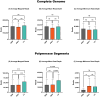
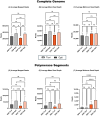
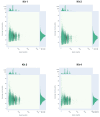


References
-
- Alnaji F. G., Holmes J. R., Rendon G., Vera J. C., Fields C. J., Martin B. E., et al. . (2019). Sequencing framework for the sensitive detection and precise mapping of defective interfering particle-associated deletions across influenza A and B viruses. J. Virol. 93 (11), e00354-19. doi: 10.1128/JVI.00354-19 - DOI - PMC - PubMed
-
- Andrés C., Del Cuerpo M., Rabella N., Piñana M., Iglesias-Cabezas M. J., González-Sánchez A., et al. . (2023). Detection of reassortant influenza B strains from 2004 to 2015 seasons in Barcelona (Catalonia, Spain) by whole genome sequencing. Virus Res. 330, 199089. doi: 10.1016/j.virusres.2023.199089 - DOI - PMC - PubMed
-
- Andrews S. (2023). FastQC A quality control tool for high throughput sequence data. Babraham Bioinformatics. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

