Phosphorylation of Optineurin by protein kinase D regulates Parkin-dependent mitophagy
- PMID: 39669425
- PMCID: PMC11634986
- DOI: 10.1016/j.isci.2024.111384
Phosphorylation of Optineurin by protein kinase D regulates Parkin-dependent mitophagy
Abstract
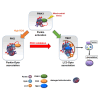
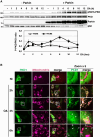
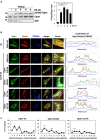
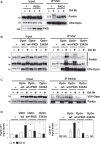
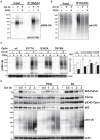
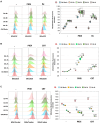
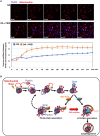
Degradation of damaged mitochondria, a process called mitophagy, plays a role in mitochondrial quality control and its dysfunction has been linked to neurodegenerative pathologies. The PINK1 kinase and the ubiquitin ligase Parkin-mediated mitophagy represents the most common pathway in which specific receptors, including Optineurin (Optn), target ubiquitin-labeled mitochondria to autophagosomes. Here, we show that Protein Kinases D (PKD) are activated and recruited to damaged mitochondria. Subsequently, PKD phosphorylate Optn to promote a complex with Parkin leading to enhancement of its ubiquitin ligase activity. Paradoxically, inhibiting PKD activity enhances the interaction between Optn and LC3, promotes the recruitment of Parkin to mitochondria, and increases the mitophagic function of Optn. This enhancement of mitophagy is characterized by increased production of mitochondrial ROS and a reduction in mitochondrial mass. The PKD kinases may therefore regulate Optn-dependent mitophagy by amplifying the Parkin-mediated degradation signals to improve the cell response against oxidative stress damage.
Keywords: Cell biology; Molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

