Integrated immunogenomic analyses of high-grade serous ovarian cancer reveal vulnerability to combination immunotherapy
- PMID: 39669575
- PMCID: PMC11634877
- DOI: 10.3389/fimmu.2024.1489235
Integrated immunogenomic analyses of high-grade serous ovarian cancer reveal vulnerability to combination immunotherapy
Abstract
Background: The efficacy of immunotherapies in high-grade serous ovarian cancer (HGSOC) is limited, but clinical trials investigating the potential of combination immunotherapy including poly-ADP-ribose polymerase inhibitors (PARPis) are ongoing. Homologous recombination repair deficiency or BRCAness and the composition of the tumor microenvironment appear to play a critical role in determining the therapeutic response.
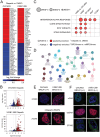
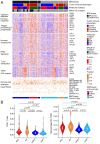
Methods: We conducted comprehensive immunogenomic analyses of HGSOC using data from several patient cohorts. Machine learning methods were used to develop a classification model for BRCAness from gene expression data. Integrated analysis of bulk and single-cell RNA sequencing data was used to delineate the tumor immune microenvironment and was validated by immunohistochemistry. The impact of PARPi and BRCA1 mutations on the activation of immune-related pathways was studied using ovarian cancer cell lines, RNA sequencing, and immunofluorescence analysis.
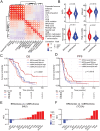
Results: We identified a 24-gene signature that predicts BRCAness. Comprehensive immunogenomic analyses across patient cohorts identified samples with BRCAness and high immune infiltration. Further characterization of these samples revealed increased infiltration of immunosuppressive cells, including tumor-associated macrophages expressing TREM2, C1QA, and LILRB4, as specified by single-cell RNA sequencing data and gene expression analysis of samples from patients receiving combination therapy with PARPi and anti-PD-1. Our findings show also that genomic instability and PARPi activated the cGAS-STING signaling pathway in vitro and the downstream innate immune response in a similar manner to HGSOC patients with BRCAness status. Finally, we have developed a web application (https://ovrseq.icbi.at) and an associated R package OvRSeq, which allow for comprehensive characterization of ovarian cancer patient samples and assessment of a vulnerability score that enables stratification of patients to predict response to the combination immunotherapy.
Conclusions: Genomic instability in HGSOC affects the tumor immune environment, and TAMs play a crucial role in modulating the immune response. Based on various datasets, we have developed a diagnostic application that uses RNA sequencing data not only to comprehensively characterize HGSOC but also to predict vulnerability and response to combination immunotherapy.
Keywords: BRCAness; PARP inhibitor; high-grade serous ovarian cancer; immunotherapy; precision oncology; tumor immune microenvironment; tumor-associated macrophages.
Copyright © 2024 Gronauer, Madersbacher, Monfort-Lanzas, Floriani, Sprung, Zeimet, Marth, Fiegl and Hackl.
Conflict of interest statement
SS was employed by Innpath GmbH. AZ reports consulting fees from Amgen, Astra Zeneca, GSK, MSD, Novartis, PharmaMar, Roche, Seagen; honoraria from Amgen, Astra Zeneca, GSK, MSD, Novartis, PharmaMar, Roche, Seagen; travel expenses from Astra Zeneca, Gilead, Roche; participation on advisory boards from Amgen, Astra Zeneca, GSK, MSD, Novartis, Pfizer, PharmaMar, Roche, Seagen. CM reports consulting fees and honoraria from Roche, Novartis, Amgen, MSD, PharmaMar, Astra Zeneca, GSK, Seagen; travel expenses from Roche, Astra Zeneca; participation on advisory boards from Roche, Novartis, Amgen, MSD, Astra Zeneca, Pfizer, PharmaMar, GSK, Seagen. HH has received research funding via Catalym and Secarna. The remaining authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

