AgoArmet and AgoC002: key effector proteins in cotton aphids host adaptation
- PMID: 39670273
- PMCID: PMC11634620
- DOI: 10.3389/fpls.2024.1500834
AgoArmet and AgoC002: key effector proteins in cotton aphids host adaptation
Abstract
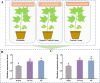
Aphids are insects that feed on phloem and introduce effector proteins into plant cells through saliva. These effector proteins are key in regulating host plant defense and enhancing aphid host adaptation. We identified these salivary proteins in the cotton aphids genome and named them AgoArmet and AgoC002. Multiple sequence alignment, protein structure analysis, and phylogenetic analysis of these proteins with related proteins from other insects showed that AgoArmet and Armet of Aphis craccivora have high sequence identity (97%) and belong to the same evolutionary branch and that AgoC002 shares the highest sequence identity (80%) and closest evolutionary relationship with C002 of Aphis glyceins. Expression profiling of AgoArmet and AgoC002 showed that they were most highly expressed in cotton aphids during the adult-3d period. Cotton aphids transferred to zucchini leaves resulted in a significant increase in the expression of AgoArmet and AgoC002 within 48h. To investigate the functions of AgoArmet and AgoC002, we decreased the expression of these genes in cotton using virus-induced gene silencing (VIGS), which ultimately led to a 38% and 26% decrease in cotton aphids fecundity, respectively. Moreover, the reduction in AgoC002 expression resulted in a significant (24%) reduction in body weight. Taken together, our findings demonstrate that AgoArmet and AgoC002 are key effector proteins involved in cotton aphids feeding and host adaptation.
Keywords: Armet; C002; VIGS; aphid; plant defense; salivary protein.
Copyright © 2024 Xue, Yan, Zhu, Wang, Chen, Luo, Cui and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

