Characterization and Inhibition of the Chaperone Function of Plasmodium falciparum Glucose-Regulated Protein 94 kDa (Pf Grp94)
- PMID: 39670568
- PMCID: PMC11968560
- DOI: 10.1002/prot.26779
Characterization and Inhibition of the Chaperone Function of Plasmodium falciparum Glucose-Regulated Protein 94 kDa (Pf Grp94)
Abstract
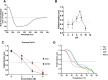
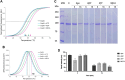
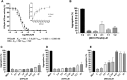
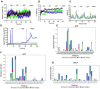
Plasmodium falciparum expresses four heat shock protein 90 (Hsp90) members. Among these, one, glucose-regulated protein 94 (PfGrp94), is localized in the endoplasmic reticulum (ER). Both the cytosolic and ER-based Hsp90s are essential for parasite survival under all growth conditions. The cytosolic version has been extensively studied and has been targeted in several efforts through the repurposing of anticancer therapeutics as antimalarial drugs. However, PfGrp94 has not been fully characterized and some of its functions related to the ER stress response are not fully understood. Structural analysis of the recombinant full-length PfGrp94 protein showed a predominantly α-helical secondary structure and its thermal resilience was modulated by 5'-N-ethyl-carboxamide-adenosine (NECA) and nucleotides ATP/ADP. PfGrp94 exhibits ATPase activity and suppressed heat-induced aggregation of a model substrate, malate dehydrogenase, in a nucleotide-dependent manner. However, these PfGrp94 chaperone functions were abrogated by NECA. Molecular docking and molecular dynamics (MD) simulations showed that NECA interacted with unique residues on PfGrp94, which could be potentially exploited for selective drug design. Finally, using parasites maintained at the red blood stage, NECA exhibited moderate antiplasmodial activity (IC50 of 4.3, 7.4, and 10.0 μM) against three different P. falciparum strains. Findings from this study provide the first direct evidence for the correlation between in silico, biochemical, and in vitro data toward utilizing the ER-based chaperone, PfGrp94, as a drug target against the malaria parasites.
Keywords: Plasmodium falciparum; NECA; characterization; endoplasmin; glucose regulated protein 94 (Grp94); inhibition; malaria.
© 2024 The Author(s). PROTEINS: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Tilley L., Dixon M. W., and Kirk K., “The Plasmodium falciparum‐Infected Red Blood Cell,” International Journal of Biochemistry & Cell Biology 43, no. 6 (2011): 839–842. - PubMed
-
- Richter K., Haslbeck M., and Buchner J., “The Heat Shock Response: Life on the Verge of Death,” Molecular Cell 40, no. 2 (2010): 253–266. - PubMed
MeSH terms
Substances
Grants and funding
- National Research Foundation South Africa
- Alexander von Humboldt Foundation, Germany
- 129401/Department of Science and Technology/National Research Foundation (NRF) of South Africa
- 145405/Department of Science and Technology/National Research Foundation (NRF) of South Africa
- CPRR23032387146/Department of Science and Technology/National Research Foundation (NRF) of South Africa
LinkOut - more resources
Full Text Sources
Miscellaneous

