Altered X-chromosome inactivation of the TLR7/8 locus and heterogeneity of pDCs in systemic sclerosis
- PMID: 39670995
- PMCID: PMC11639950
- DOI: 10.1084/jem.20231809
Altered X-chromosome inactivation of the TLR7/8 locus and heterogeneity of pDCs in systemic sclerosis
Abstract
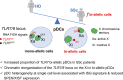
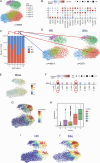
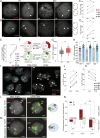
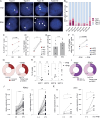
Systemic sclerosis (SSc) is an autoimmune disease that has a strong female predominance. Both the X-linked TLR7 and TLR8 can induce type I IFN (IFN-I) by plasmacytoid DCs (pDCs), which can promote fibrosis. We identified five subclusters of pDCs, including ISGhigh clusters that were over-represented in SSc patients. We observed that both TLR7 and TLR8 genes escape from X chromosome inactivation (XCI) at higher frequency in pDCs of SSc patients, which was associated with changes in TLR7 protein profile. Combined DNA/RNA FISH analysis revealed that the TLR7/8 locus is preferentially located outside of the inactive X (Xi) territory when TLR7 is expressed, suggesting that higher-order loop formation is linked to TLR7/8 expression from the Xi. Furthermore, the expression levels of XIST and the transcriptional repressor SPEN were reduced in SSc pDCs. Hence, our data revealed the heterogeneity of pDCs in SSc and suggested that altered XCI at the TLR7/8 locus may contribute to the chronic IFN-I activity of pDCs in female SSc patients.
© 2024 Du et al.
Conflict of interest statement
Disclosures: R.F. Spiera reported personal fees from Chemocentryx, AbbVie/Abbott, BMS, Regeneron,Amgen, Vera, GlaxoSmithKline, Novartis, Boehringer Ingelheim, Galderma, Sanofi, Roche Genentech, Cytori, and Astra-Zeneca, and grants from Horizon Pharma, AbbVie/Abbott, AstraZeneca, ChemoCentryx, Corbus, Genentech, GlaxoSmithKline, InflaRx, Kadmon, Novartis, Principia, Roche, Sanofi, Cytori, Boehringer Ingelheim, and Amgen outside the submitted work. M.-E. Truchetet reported personal fees from Abbvie, Boehringer, Janssen, Lilly, and Astrazeneca outside the submitted work. F.J. Barrat reported “other” from Ipinovyx Bio and Epistemyx Bio outside the submitted work. No other disclosures were reported.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

