Asparagine availability controls germinal center B cell homeostasis
- PMID: 39671468
- PMCID: PMC7617476
- DOI: 10.1126/sciimmunol.adl4613
Asparagine availability controls germinal center B cell homeostasis
Abstract
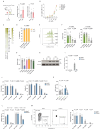
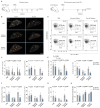
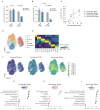
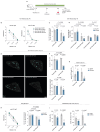
The rapid proliferation of germinal center (GC) B cells requires metabolic reprogramming to meet energy demands, yet these metabolic processes are poorly understood. By integrating metabolomic and transcriptomic profiling of GC B cells, we identified that asparagine (Asn) metabolism was highly up-regulated and essential for B cell function. Asparagine synthetase (ASNS) was up-regulated after B cell activation through the integrated stress response sensor GCN2. Conditional deletion of Asns in B cells impaired survival and proliferation in low Asn conditions. Removal of environmental Asn by asparaginase or dietary restriction compromised the GC reaction, impairing affinity maturation and the humoral response to influenza infection. Furthermore, metabolic adaptation to the absence of Asn required ASNS, and oxidative phosphorylation, mitochondrial homeostasis, and synthesis of nucleotides were particularly sensitive to Asn deprivation. These findings demonstrate that Asn metabolism acts as a key regulator of B cell function and GC homeostasis.
Conflict of interest statement
The authors have no competing interests.
Figures




References
-
- Victora D, Nussenzweig MC. Germinal Centers. Annu Rev Immunol. 2022;40:413–442. - PubMed
-
- Chen D, Wang Y, Manakkat Vijay GK, Fu S, Nash CW, Xu D, He D, Salomonis N, Singh H, Xu H. Coupled Analysis of Transcriptome and BCR Mutations Reveals Role of OXPHOS in Affinity Maturation. Nat Immunol. 2021;22:904–913. - PubMed
-
- Weisel FJ, Mullett SJ, Elsner RA, Menk AV, Trivedi N, Luo W, Wikenheiser D, Hawse WF, Chikina M, Smita S, Conter LJ, et al. Germinal Center B Cells Selectively Oxidize Fatty Acids for Energy While Conducting Minimal Glycolysis. Nat Immunol. 2020;21:331–342. doi: 10.1038/s41590-020-0598-4. - DOI - PMC - PubMed
-
- Urbanczyk S, Baris OR, Hofmann J, Taudte RV, Guegen N, Golombek F, Castiglione K, Meng X, Bozec A, Thomas J, Weckwerth L, et al. Mitochondrial Respiration in B Lymphocytes Is Essential for Humoral Immunity by Controlling the Flux of the TCA Cycle. Cell Rep. 2022;39:110912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

