Targeted protein degradation of PDE4 shortforms by a novel proteolysis targeting chimera
- PMID: 39673076
- PMCID: PMC12220845
- DOI: 10.1111/febs.17359
Targeted protein degradation of PDE4 shortforms by a novel proteolysis targeting chimera
Abstract
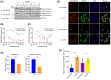
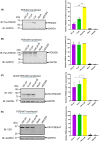
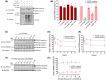
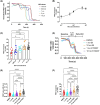
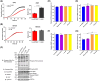
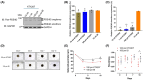
Cyclic AMP (cAMP) has a crucial role in many vital cellular processes and there has been much effort expended in the discovery of inhibitors against the enzyme superfamily that degrades this second messenger, namely phosphodiesterases (PDEs). The journey of competitive PDE inhibitors to the clinic has been hampered by side effects profiles that have resulted from a lack of selectivity for subfamilies and individual isoforms because of high conservation of catalytic site sequences and structures. Here we introduce a proteolysis targeting chimera (PROTAC) that can specifically target a small subset of isoforms from the PDE4 family to send the enzyme for degradation at the proteasome by recruiting a ubiquitin E3 ligase into proximity with the PDE. We constructed our PDE4 PROTAC (KTX207) using a previously characterized PDE4 inhibitor, and we show that evolution of the compound into a PROTAC improves selectivity, potency and enables a long-lasting effect even after the compound is removed from cells after a short treatment duration. Functionally, KTX207 is more effective at increasing cAMP, is 100 times more anti-inflammatory, and is significantly better at reducing the growth in cancer cell models than the PDE4 inhibitor alone. Our study highlights the advantages of targeted degradation versus active-site occupancy for PDE4 inhibition and discusses the potential of this novel pharmacological approach to improve the safety profile of PDE4 inhibition in the future.
Keywords: PDE4; PROTAC; phosphodiesterases; protein degradation.
© 2024 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
CS, KC, and DS are founders of Katalytic Therapeutics and hold patent rights related to KTX207. The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

