Hypusination in intestinal epithelial cells protects mice from infectious colitis
- PMID: 39673545
- PMCID: PMC11649231
- DOI: 10.1080/19490976.2024.2438828
Hypusination in intestinal epithelial cells protects mice from infectious colitis
Abstract
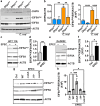
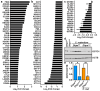
Enteropathogenic Escherichia coli (EPEC) is a bacterium that causes attaching/effacing (A/E) lesions and serious diarrheal disease, a major health issue in developing countries. EPEC pathogenicity results from the effect of virulence factors and dysregulation of host responses. Polyamines, including spermidine, play a major role in intestinal homeostasis. Spermidine is the substrate for deoxyhypusine synthase (DHPS), which catalyzes the conjugation of the amino acid hypusine to eukaryotic translation initiation factor 5A (EIF5A); hypusinated EIF5A (EIF5AHyp) binds specific mRNAs and initiates translation. Our aim was to determine the role of hypusination during infection with A/E pathogens. We found that DHPS and EIF5AHyp levels are induced in i) a colonic epithelial cell line and human-derived colon organoids infected with EPEC, and ii) the colon of mice infected with Citrobacter rodentium, the rodent equivalent of EPEC. Specific deletion of Dhps in intestinal epithelial cells worsened clinical, histological, and pro-inflammatory parameters in C. rodentium-infected mice. These animals also exhibited an exacerbated pathogenic transcriptome in their colon. Furthermore, infected mice with specific Dhps deletion exhibited reduced levels of proteins involved in detoxification of tissue-damaging reactive aldehydes and consequently increased electrophile adducts in the colon. Thus, hypusination in intestinal epithelial cells protects from infectious colitis mediated by A/E pathogens.
Keywords: Citrobacter rodentium; Enteropathogenic Escherichia coli; Polyamines; attaching and effacing pathogen; colitis; host–pathogen interactions; hypusine; mucosal immune response; translation.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





References
-
- Lanata CF, Fischer-Walker CL, Olascoaga AC, Torres CX, Torres MJ, Black, RE. Child Health Epidemiology Reference Group of the World Health Organization and Unicef . Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLOS ONE. 2013;8(9):e72788. doi:10.1371/journal.pone.0072788. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
