Identification of a distinctive immunogenomic gene signature in stage-matched colorectal cancer
- PMID: 39673574
- PMCID: PMC11646222
- DOI: 10.1007/s00432-024-06034-4
Identification of a distinctive immunogenomic gene signature in stage-matched colorectal cancer
Abstract
Background: Colorectal cancer (CRC) remains one of the leading causes of cancer-related mortality worldwide. Despite advances in diagnosis and treatment, including surgery, chemotherapy, and immunotherapy, accurate clinical markers are still lacking. The development of prognostic and predictive indicators, particularly in the context of personalized medicine, could significantly improve CRC patient management.
Method: In this retrospective study, we used FFPE blocks of tissue samples from CRC patients at Augusta University (AU) to quantify a custom 15-gene panel. To differentiate the tumor and adjacent normal regions (NAT), H&E staining was utilized. For the quantification of transcripts, we used the NanoString nCounter platform. Kaplan-Meier and Log-rank tests were used to perform survival analyses. Several independent datasets were explored to validate the gene signature. Orthogonal analyses included single-cell profiling, differential gene expression, immune cell deconvolution, neoantigen prediction, and biological pathway assessment.
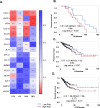
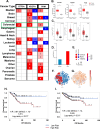
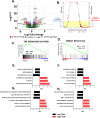
Results: A 3-gene signature (GTF3A, PKM, and VEGFA) was found to be associated with overall survival in the AU cohort (HR = 2.26, 95% CI 1.05-4.84, p = 0.02, 93 patients), TCGA cohort (HR = 1.57, 95% CI 1.05-2.35, p < 0.02, 435 patients) and four other GEO datasets. Independent single-cell analysis identified relatively higher expression of the 3-gene signature in the tumor region. Differential analysis revealed dysregulated tissue inflammation, immune dysfunction, and neoantigen load of cell cycle processes among high-risk patients compared to low-risk patients.
Conclusion: We developed a 3-gene signature with the potential for prognostic and predictive clinical assessment of CRC patients. This gene-based stratification offers a cost-effective approach to personalized cancer management. Further research using similar methods could identify therapy-specific gene signatures to strengthen the development of personalized medicine for CRC patients.
Keywords: Colon; Colorectal cancer; Gene signature; Immune infiltration; Immunotherapy responsiveness; Personalized medicine; Precision medicine; Prognostic genes; Stratified medicine.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflcit of interest: The authors declare no competing interests. Ethical Standards: The study was approved by the Institutional Review Board (IRB-HAC # 611298) of Augusta University. Consent to participate: The research was conducted using de-identified samples and no consent was required. Consent for publication: N/A.
Figures



References
-
- Anuraga G, Tang WC, Phan NN, Ta HDK, Liu YH, Wu YF, Wang CY (2021) Comprehensive analysis of prognostic and genetic signatures for general transcription factor III (GTF3) in clinical colorectal cancer patients using bioinformatics approaches. Curr Issues Mol Biol, 43(1): 2–20. 10.3390/cimb43010002 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

