Lig3-dependent rescue of mouse viability and DNA double-strand break repair by catalytically inactive Lig4
- PMID: 39673806
- PMCID: PMC11754673
- DOI: 10.1093/nar/gkae1216
Lig3-dependent rescue of mouse viability and DNA double-strand break repair by catalytically inactive Lig4
Abstract
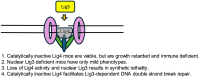
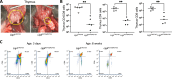
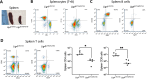
Recent studies have revealed a structural role for DNA ligase 4 (Lig4) in the maintenance of a repair complex during non-homologous end joining (NHEJ) of DNA double-strand breaks. In cultured cell lines, catalytically inactive Lig4 can partially alleviate the severe DNA repair phenotypes observed in cells lacking Lig4. To study the structural role of Lig4 in vivo, a mouse strain harboring a point mutation to Lig4's catalytic site was generated. In contrast to the ablation of Lig4, catalytically inactive Lig4 mice are born alive. These mice display marked growth retardation and have clear deficits in lymphocyte development. We considered that the milder phenotype results from inactive Lig4 help to recruit another ligase to the repair complex. We next generated a mouse strain deficient for nuclear Lig3. Nuclear Lig3-deficient mice are moderately smaller and have elevated incidences of cerebral ventricle dilation but otherwise appear normal. Strikingly, in experiments crossing these two strains, mice lacking nuclear Lig3 and expressing inactive Lig4 were not obtained. Timed mating revealed that fetuses harboring both mutations underwent resorption, establishing an embryonic lethal genetic interaction. These data suggest that Lig3 is recruited to NHEJ complexes to facilitate end joining in the presence (but not activity) of Lig4.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Joining of DNA breaks- interplay between DNA ligases and poly (ADP-ribose) polymerases.DNA Repair (Amst). 2025 May;149:103843. doi: 10.1016/j.dnarep.2025.103843. Epub 2025 May 2. DNA Repair (Amst). 2025. PMID: 40347914 Review.
-
The model moss Physcomitrium patens relies heavily on homologous recombination to repair DNA double-strand breaks.DNA Repair (Amst). 2025 Aug;152:103881. doi: 10.1016/j.dnarep.2025.103881. Epub 2025 Aug 5. DNA Repair (Amst). 2025. PMID: 40803178
-
PIPKIγ promotes non-homologous end joining through LIG4 to enhance radiotherapy resistance in triple-negative breast cancer.Cell Death Dis. 2025 Jul 31;16(1):578. doi: 10.1038/s41419-025-07894-5. Cell Death Dis. 2025. PMID: 40744919 Free PMC article.
-
Dynamic assemblies and coordinated reactions of non-homologous end joining.Nature. 2025 Jul;643(8072):847-854. doi: 10.1038/s41586-025-09078-9. Epub 2025 Jun 11. Nature. 2025. PMID: 40500445
-
DNA-PK: A synopsis beyond synapsis.DNA Repair (Amst). 2024 Sep;141:103716. doi: 10.1016/j.dnarep.2024.103716. Epub 2024 Jul 8. DNA Repair (Amst). 2024. PMID: 38996771 Review.
Cited by
-
Joining of DNA breaks- interplay between DNA ligases and poly (ADP-ribose) polymerases.DNA Repair (Amst). 2025 May;149:103843. doi: 10.1016/j.dnarep.2025.103843. Epub 2025 May 2. DNA Repair (Amst). 2025. PMID: 40347914 Review.
-
Reversal gene expression assessment for drug repurposing, a case study of glioblastoma.J Transl Med. 2025 Jan 7;23(1):25. doi: 10.1186/s12967-024-06046-1. J Transl Med. 2025. PMID: 39773231 Free PMC article.
References
-
- Levin D.S., McKenna A.E., Motycka T.A., Matsumoto Y., Tomkinson A.E.. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000; 10:919–922. - PubMed
-
- Montecucco A., Rossi R., Levin D.S., Gary R., Park M.S., Motycka T.A., Ciarrocchi G., Villa A., Biamonti G., Tomkinson A.E.. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998; 17:3786–3795. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

