Insights into the human cDNA: A descriptive study using library screening in yeast
- PMID: 39674632
- PMCID: PMC11533663
- DOI: 10.1016/j.jgeb.2024.100427
Insights into the human cDNA: A descriptive study using library screening in yeast
Abstract
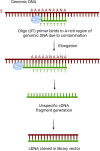
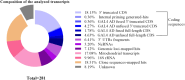
The utilization of human cDNA libraries in yeast genetic screens is an approach that has been used to identify novel gene functions and/or genetic and physical interaction partners through forward genetics using yeast two-hybrid (Y2H) and classical cDNA library screens. Here, we summarize several challenges that have been observed during the implementation of human cDNA library screens in Saccharomyces cerevisiae (budding yeast). Upon the utilization of DNA repair deficient-yeast strains to identify novel genes that rescue the toxic effect of DNA-damage inducing drugs, we have observed a wide range of transcripts that could rescue the strains. However, after several rounds of screening, most of these hits turned out to be false positives, most likely due to spontaneous mutations in the yeast strains that arise as a rescue mechanism due to exposure to toxic DNA damage inducing-drugs. The observed transcripts included mitochondrial hits, non-coding RNAs, truncated cDNAs, and transcription products that resulted from the internal priming of genomic regions. We have also noticed that most cDNA transcripts are not fused with the GAL4 activation domain (GAL4AD), rendering them unsuitable for Y2H screening. Consequently, we utilized Sanger sequencing to screen 282 transcripts obtained from either four different yeast screens or through direct fishing from a human kidney cDNA library. The aim was to gain insights into the different transcription products and to highlight the challenges of cDNA screening approaches in the presence of a significant number of undesired transcription products. In summary, this study describes the challenges encountering human cDNA library screening in yeast as a valuable technique that led to the identification of important molecular mechanisms. The results open research venues to further optimize the process and increase its efficiency.
Keywords: Genetic screens; Saccharomyces cerevisiae; Yeast two-hybrid; cDNA library screens.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Banik S.S.R., Counter C.M. From bread to bedside: what budding yeast has taught us about the immortalization of cancer cells. Yeast as a Tool Cancer Res. 2007:123–139. doi: 10.1007/978-1-4020-5963-6_5. - DOI
-
- Nam D.K., Lee S., Zhou G., et al. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. PNAS. 2002;99:6152–6156. doi: 10.1073/PNAS.092140899/ASSET/08839A23-CD85-4252-8E76-5928DB218856/ASSETS/GRAPHIC/PQ0921408005.JPEG. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
