Sarcopenic obesity is attenuated by E-syt1 inhibition via improving skeletal muscle mitochondrial function
- PMID: 39675068
- PMCID: PMC11699297
- DOI: 10.1016/j.redox.2024.103467
Sarcopenic obesity is attenuated by E-syt1 inhibition via improving skeletal muscle mitochondrial function
Abstract
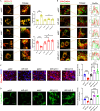
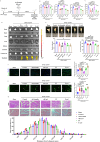
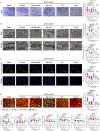
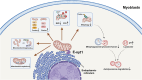
In aging and metabolic disease, sarcopenic obesity (SO) correlates with intramuscular adipose tissue (IMAT). Using bioinformatics analysis, we found a potential target protein Extended Synaptotagmin 1 (E-syt1) in SO. To investigate the regulatory role of E-syt1 in muscle metabolism, we performed in vivo and in vitro experiments through E-syt1 loss- and gain-of-function on muscle physiology. When E-syt1 is overexpressed in vitro, myoblast proliferation, differentiation, mitochondrial respiration, biogenesis, and mitochondrial dynamics are impaired, which were alleviated by the silence of E-syt1. Furthermore, overexpression of E-syt1 inhibited mitophagic flux. Mechanistically, E-syt1 overexpression leads to mitochondrial calcium overload and mitochondrial ROS burst, inhibits the fusion of mitophagosomes with lysosomes, and impedes the acidification of lysosomes. Animal experiments demonstrated the inhibition of E-syt1 increased the capacity of endurance exercise, muscle mass, mitochondrial function, and oxidative capacity of the muscle fibers in OVX mice. These findings establish E-syt1 as a novel contributor to the pathogenesis of skeletal muscle metabolic disorders in SO. Consequently, targeting E-syt1-induced dysfunction may serve as a viable strategy for attenuating SO.
Keywords: E-syt1; Mitochondria; Mitophagy; Myogenesis; Sarcopenic obesity.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

