AMPK protects proximal tubular epithelial cells from lysosomal dysfunction and dedifferentiation induced by lipotoxicity
- PMID: 39675352
- PMCID: PMC11925112
- DOI: 10.1080/15548627.2024.2435238
AMPK protects proximal tubular epithelial cells from lysosomal dysfunction and dedifferentiation induced by lipotoxicity
Abstract
Renal proximal tubules are a primary site of injury in metabolic diseases. In obese patients and animal models, proximal tubular epithelial cells (PTECs) display dysregulated lipid metabolism, organelle dysfunctions, and oxidative stress that contribute to interstitial inflammation, fibrosis and ultimately end-stage renal failure. Our research group previously pointed out AMP-activated protein kinase (AMPK) decline as a driver of obesity-induced renal disease. Because PTECs display high macroautophagic/autophagic activity and rely heavily on their endo-lysosomal system, we investigated the effect of lipid stress on autophagic flux and lysosomes in these cells. Using a model of highly differentiated primary PTECs challenged with palmitate, our data placed lysosomes at the cornerstone of the lipotoxic phenotype. As soon as 6 h after palmitate exposure, cells displayed impaired lysosomal acidification subsequently leading to autophagosome accumulation and activation of lysosomal biogenesis. We also showed the inability of lysosomal quality control to restore acidic pH which finally drove PTECs dedifferentiation. When palmitate-induced AMPK activity decline was prevented by AMPK activators, lysosomal acidification and the differentiation profile of PTECs were preserved. Our work provided key insights on the importance of lysosomes in PTECs homeostasis and lipotoxicity and demonstrated the potential of AMPK in protecting the organelle from lipid stress.Abbreviation: ACAC: acetyl-CoA carboxylase; ACTB: actin beta; AICAR: 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside; AMPK: AMP-activated protein kinase; APQ1: aquaporin 1 (Colton blood group); BSA: bovine serum albumin; CDH16: cadherin 16; CKD: chronic kidney disease; CTSB: cathepsin B; CTSD: cathepsin D; EPB41L5: erythrocyte membrane protein band 4.1 like 5; EIF4EBP1: eukaryotic translation initiation factor 4E binding protein 1; EMT: epithelial-to-mesenchymal transition; FA: fatty acid; FCCP: carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; GFP: green fluorescent protein; GUSB: glucuronidase beta; HEXB: hexosaminidase subunit beta; LAMP: lysosomal associated membrane protein; LD: lipid droplet; LGALS3: galectin 3; LLOMe: L-leucyl-L-leucine methyl ester hydrobromide; LMP: lysosomal membrane permeabilization; LRP2: LDL receptor related protein 2; LSD: lysosomal storage disorder; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MCOLN1: mucolipin TRP cation channel 1; MG132: N-benzyloxycarbonyl-L-leucyl-L-leucyl-L-leucinal; MmPTECs: Mus musculus (mouse) proximal tubular epithelial cells; MTORC1: mechanistic target of rapamycin kinase complex 1; OA: oleate; PA: palmitate; PIKFYVE: phosphoinositide kinase, FYVE-type zinc finger containing; PTs: proximal tubules; PTECs: proximal tubular epithelial cells; PRKAA: protein kinase AMP-activated catalytic subunit alpha; RFP: red fluorescent protein; RPS6KB: ribosomal protein S6 kinase B; SLC5A2: solute carrier family 5 member 2; SOX9: SRY-box transcription factor 9; SQSTM1: sequestosome 1; TFEB: transcription factor EB; Ub: ubiquitin; ULK1: unc-51 like autophagy activating kinase 1; VIM: vimentin.
Keywords: AMPK; Autophagy; chronic kidney disease; lipid accumulation; obesity; proximal tubules.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures








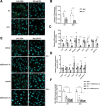
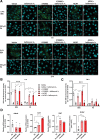
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
