Redirecting E3 ubiquitin ligases for targeted protein degradation with heterologous recognition domains
- PMID: 39675716
- PMCID: PMC11758572
- DOI: 10.1016/j.jbc.2024.108077
Redirecting E3 ubiquitin ligases for targeted protein degradation with heterologous recognition domains
Abstract
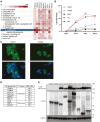
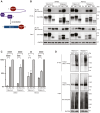
Targeted protein degradation (TPD) mediated by proteolysis targeting chimeras or molecular glues is an emerging therapeutic strategy. Despite greater than 600 E3 ligases and their associated components, a limited number have been deployed in TPD. Those commonly used include cereblon and von Hippel-Lindau tumor suppressor (VHL), which is expressed widely and for which high affinity ligands are available. Limiting TPD to specific cells or tissues would be desirable in many settings. To this goal we have determined the potential of two erythroid cell-enriched E3 ligases, TRIM10 and TRIM58, to degrade a protein of interest, BCL11A, a validated therapeutic target for the β-hemoglobinopathies. We established a general strategy in which heterologous recognition domains replace the PRY-SPRY domain of TRIM10 and TRIM58. Recruitment of TRIM10 or TRIM58 to BCL11A by coiled-coil peptides, nanobodies, or the substrate recognition domain of cereblon led to its degradation. Our findings illustrate a strategy that may be widely useful in evaluating the TPD potential of other E3 ubiquitin ligases and provide a rationale for discovery of ligands for TRIM10 and TRIM58 for erythroid-selective depletion of proteins of interest.
Keywords: BCL11A; E3 ubiquitin ligases; PROTACs; TRIM10; TRIM58; targeted protein degradation.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest S. H. O. is an Investigator of the Howard Hughes Medical Institute. The other authors declare that they had no conflicts of interest with the contents of this article.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

