Conditional Overexpression of Net1 Enhances the Trans-Differentiation of Lgr5+ Progenitors into Hair Cells in the Neonatal Mouse Cochlea
- PMID: 39675772
- PMCID: PMC11969244
- DOI: 10.1111/cpr.13787
Conditional Overexpression of Net1 Enhances the Trans-Differentiation of Lgr5+ Progenitors into Hair Cells in the Neonatal Mouse Cochlea
Abstract
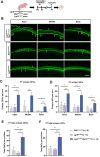
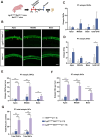
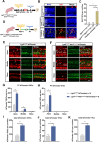
Sensorineural hearing loss is mainly caused by damage to hair cells (HC), which cannot be regenerated spontaneously in adult mammals once damaged. Cochlear Lgr5+ progenitors are characterised by HC regeneration capacity in neonatal mice, and we previously screened several new genes that might induce HC regeneration from Lgr5+ progenitors. Net1, a guanine nucleotide exchange factor, is one of the screened new genes and is particularly active in cancer cells and is involved in cell proliferation and differentiation. Here, to explore in vivo roles of Net1 in HC regeneration, Net1 loxp/loxp mice were constructed and crossed with Lgr5 CreER/+ mice to conditionally overexpress (cOE) Net1 in cochlear Lgr5+ progenitors. We observed a large number of ectopic HCs in Lgr5 CreER/+ Net1 loxp/loxp mouse cochlea, which showed a dose-dependent effect. Moreover, the EdU assay was unable to detect any EdU+/Sox2+ supporting cells, while lineage tracing showed significantly more regenerated tdTomato+ HCs in Lgr5 CreER/+ Net1 loxp/loxp tdTomato mice, which indicated that Net1 cOE enhanced HC regeneration by inducing the direct trans-differentiation of Lgr5+ progenitors rather than mitotic HC regeneration. Additionally, qPCR results showed that the transcription factors related to HC regeneration, including Atoh1, Gfi1 and Pou4f3, were significantly upregulated and are probably the mechanism behind the HC regeneration induced by Net1. In conclusion, our study provides new evidence for the role of Net1 in enhancing HC regeneration in the neonatal mouse cochlea.
Keywords: Net1; HC regeneration; Lgr5+ progenitors; proliferation; trans‐differentiation.
© 2024 The Author(s). Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Kelley M. W., “Regulation of Cell Fate in the Sensory Epithelia of the Inner Ear,” Nature Reviews Neuroscience 7, no. 11 (2006): 837–849. - PubMed
-
- Jang M. W., Lim J., Park M. G., Lee J. H., and Lee C. J., “Active Role of Glia‐Like Supporting Cells in the Organ of Corti: Membrane Proteins and Their Roles in Hearing,” Glia 70, no. 10 (2022): 1799–1825. - PubMed
-
- Revuelta M., Santaolalla F., Arteaga O., Alvarez A., Sánchez‐del‐Rey A., and Hilario E., “Recent Advances in Cochlear Hair Cell Regeneration‐A Promising Opportunity for the Treatment of Age‐Related Hearing Loss,” Ageing Research Reviews 36 (2017): 149–155. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFA0807000/National Key Research and Development Program of China
- 2023YFA1801804/National Key Research and Development Program of China
- 2021YFA1101300/National Key Research and Development Program of China
- 2021YFA1101800/National Key Research and Development Program of China
- 2020YFA0112503/National Key Research and Development Program of China
- 82171149/National Natural Science Foundation of China
- 82371166/National Natural Science Foundation of China
- 82330033/National Natural Science Foundation of China
- 82030029/National Natural Science Foundation of China
- 81970882/National Natural Science Foundation of China
- 92149304/National Natural Science Foundation of China
- 22302231/National Natural Science Foundation of China
- JCYJ20230807114700001/Shenzhen Science and Technology Program
- JCYJ20210324125608022/Shenzhen Science and Technology Program
- JCYJ20190814093401920/Shenzhen Science and Technology Program
- JCYJ20190808120405672/Shenzhen Science and Technology Program
- 2024A1515010548/Guangdong Basic and Applied Basic Research Foundation
- 2021YFS0371/Science and Technology Department of Sichuan Province
- YKY-KF202201/Open Research Fund of Guangdong Academy of Medical Sciences
LinkOut - more resources
Full Text Sources
Medical

