ID1 and ID3 functions in the modulation of the tumour immune microenvironment in adult patients with B-cell acute lymphoblastic leukaemia
- PMID: 39676870
- PMCID: PMC11638060
- DOI: 10.3389/fimmu.2024.1473909
ID1 and ID3 functions in the modulation of the tumour immune microenvironment in adult patients with B-cell acute lymphoblastic leukaemia
Abstract
Introduction: B-cell acute lymphoblastic leukemia (B-ALL) in adults often presents a poor prognosis. ID1 and ID3 genes have been identified as predictors of poor response in Colombian adult B-ALL patients, contributing to cancer development. In various cancer models, these genes have been associated with immune regulatory populations within the tumor immune microenvironment (TIME). B-ALL progression alters immune cell composition and the bone marrow (BM) microenvironment, impacting disease progression and therapy response. This study investigates the relationship between ID1 and ID3 expression, TIME dynamics, and immune evasion mechanisms in adult B-ALL patients.
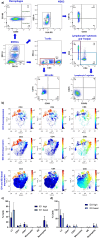
Methods: This exploratory study analysed BM samples from 10 B-ALL adult patients diagnosed at the National Cancer Institute of Colombia. First, RT-qPCR was used to assess ID1 and ID3 expression in BM tumour cells. Flow cytometry characterised immune populations in the TIME. RNA-seq evaluated immune genes associatedwith B-ALL immune response, while xCell and CytoSig analysed TIME cell profiles and cytokines. Pathway analysis, gene ontology, and differential gene expression (DEGs) were examined, with functional enrichment analysis performed using KEGG ontology.
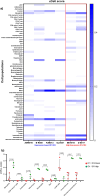
Results: Patients were divided into two groups based on ID1 and ID3 expression, namely basal and overexpression. A total of 94 differentially expressed genes were identified between these groups, with top overexpressed genes associated with neutrophil pathways. Gene set enrichment analysis revealed increased expression of genes associated with neutrophil degranulation, immune response-related neutrophil activation, and neutrophil-mediated immunity. These findings correlated with xCell data. Overexpression group showed significant differences in neutrophils, monocytes and CD4+ naive T cells compared to basal group patients. Microenvironment and immune scores were also significantly different, consistent with the flow cytometry results. Elevated cytokine levels associated with neutrophil activation supported these findings. Validation was performed using the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) TCGA B-ALL cohorts.
Discussion: These findings highlight significant differences in ID1 and ID3 expression levels and their impact on TIME populations, particularly neutrophil-related pathways. The results suggest a potential role for ID1 and ID3 in immune evasion in adult B-ALL, mediated through neutrophil activation and immune regulation.
Keywords: B-cell acute lymphoblastic leukaemia (B-ALL); bone marrow; immune evasion; immune system; immunosurveillance; microenvironment.
Copyright © 2024 Poveda-Garavito, Orozco Castaño, Torres-Llanos, Cruz-Rodriguez, Parra-Medina, Quijano, Zabaleta and Combita.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Felipe Combariza J, Casas CP, Rodriguez M. Supervivencia en adultos con leucemia linfoide aguda de novo tratados con el esquema HyperCVAD en el Instituto Nacional de Cancerología (Colombia), entre enero de 2001 y junio de 2005. Rev Colomb CanCeRol. (2007) 11(2):92–100.
-
- Chiarini F, Lonetti A, Evangelisti C, Buontempo F, Orsini E, Evangelisti C, et al. Advances in understanding the acute lymphoblastic leukemia bone marrow microenvironment: From biology to therapeutic targeting. Biochim Biophys Acta Mol Cell Res. (2016) 1863:449–63. doi: 10.1016/j.bbamcr.2015.08.015 - DOI - PubMed
-
- Xu K, Feng Q, Wiemels JL, de Smith AJ. Disparities in acute lymphoblastic leukemia risk and survival across the lifespan in the United States of America. J Transl Genet Genom. (2021) 5:218–39. https://www.oaepublish.com/articles/jtgg.2021.20.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

