This is a preprint.
Genome-wide Machine Learning Analysis of Anosmia and Ageusia with COVID-19
- PMID: 39677430
- PMCID: PMC11643161
- DOI: 10.1101/2024.12.04.24318493
Genome-wide Machine Learning Analysis of Anosmia and Ageusia with COVID-19
Abstract
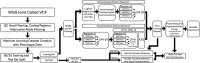
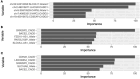
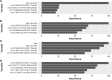
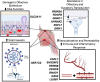
The COVID-19 pandemic has caused substantial worldwide disruptions in health, economy, and society, manifesting symptoms such as loss of smell (anosmia) and loss of taste (ageusia), that can result in prolonged sensory impairment. Establishing the host genetic etiology of anosmia and ageusia in COVID-19 will aid in the overall understanding of the sensorineural aspect of the disease and contribute to possible treatments or cures. By using human genome sequencing data from the University of Iowa (UI) COVID-19 cohort (N=187) and the National Institute of Health All of Us (AoU) Research Program COVID-19 cohort (N=947), we investigated the genetics of anosmia and/or ageusia by employing feature selection techniques to construct a novel variant and gene prioritization pipeline, utilizing machine learning methods for the classification of patients. Models were assessed using a permutation-based variable importance (PVI) strategy for final prioritization of candidate variants and genes. The highest held-out test set area under the receiver operating characteristic (AUROC) curve for models and datasets from the UI cohort was 0.735 and 0.798 for the variant and gene analysis respectively and for the AoU cohort was 0.687 for the variant analysis. Our analysis prioritized several novel and known candidate host genetic factors involved in immune response, neuronal signaling, and calcium signaling supporting previously proposed hypotheses for anosmia/ageusia in COVID-19.
Conflict of interest statement
COMPETING INTEREST STATMENT The authors declare no conflict of interest.
Figures
References
-
- Abdelhafez M, Nasereddin A, Shamma OA, Abed R, Sinnokrot R, Marof O, Heif T, Erekat Z, Al-Jawabreh A, Ereqat S. 2023. Association of IFNAR2 rs2236757 and OAS3 rs10735079 Polymorphisms with Susceptibility to COVID-19 Infection and Severity in Palestine. Interdiscip Perspect Infect Dis 2023: 9551163. doi: 10.1155/2023/9551163 - DOI - PMC - PubMed
-
- Alzubi R, Ramzan N, Alzoubi H, Amira A. 2017. A hybrid feature selection method for complex diseases SNPs. IEEE Access 6: 1292–1301. doi: 10.1109/ACCESS.2017.2778268 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
