This is a preprint.
Consultation informs strategies to improve functional evidence use in variant classification
- PMID: 39677445
- PMCID: PMC11643179
- DOI: 10.1101/2024.12.04.24318523
Consultation informs strategies to improve functional evidence use in variant classification
Update in
-
Consultation informs strategies for improving the use of functional evidence in variant classification.Am J Hum Genet. 2025 Jun 5;112(6):1489-1495. doi: 10.1016/j.ajhg.2025.05.003. Am J Hum Genet. 2025. PMID: 40480201 Free PMC article.
Abstract
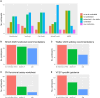
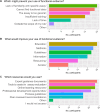
To determine if a variant identified by diagnostic genetic testing is causal for disease, applied genetics professionals evaluate all available evidence to assign a clinical classification. Experimental assay data can provide strong functional evidence for or against pathogenicity in variant classification, but appears to be underutilised. We surveyed genetic diagnostic professionals in Australasia to assess their application of functional evidence in clinical practice. Results indicated that survey respondents are not confident to apply functional evidence, mainly due to uncertainty around practice recommendations. Respondents also identified need for support resources, educational opportunities, and in particular requested expert recommendations and updated practice guidelines to improve translation of experimental data to curation evidence. As an initial step, we have collated a list of functional assays recommended by 19 ClinGen Variant Curation Expert Panels as a source of international expert opinion on functional evidence evaluation. Additional support resources for diagnostic practice are in development.
Keywords: Functional evidence; assay; diagnostic genetics; education; variant classification.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures
References
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. - PMC - PubMed
-
- Rivera-Munoz EA, Milko LV, Harrison SM, Azzariti DR, Kurtz CL, Lee K, et al. ClinGen Variant Curation Expert Panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation. Hum Mutat. 2018;39(11):1614–22. - PMC - PubMed
-
- ClinGen. Criteria Specification Registry 2024. [Available from: https://cspec.genome.network/cspec/ui/svi/.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
