This is a preprint.
Establishing methods to monitor H5N1 influenza virus in dairy cattle milk
- PMID: 39677482
- PMCID: PMC11643214
- DOI: 10.1101/2024.12.04.24318491
Establishing methods to monitor H5N1 influenza virus in dairy cattle milk
Update in
-
Establishing Methods to Monitor Influenza (A)H5N1 Virus in Dairy Cattle Milk, Massachusetts, USA.Emerg Infect Dis. 2025 May;31(13):70-75. doi: 10.3201/eid3113.250087. Epub 2025 Mar 26. Emerg Infect Dis. 2025. PMID: 40138725 Free PMC article.
Abstract
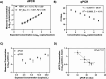
Highly Pathogenic Avian Influenza strain H5N1 has caused a multi-state outbreak among US dairy cattle, spreading across 15 states and infecting hundreds of herds since its onset. We rapidly developed and optimized PCR-based detection assays and sequencing protocols to support H5N1 molecular surveillance. Using 214 retail milk from 20 states for methods development, we found that H5N1 concentrations by digital PCR strongly correlated with qPCR cycle threshold (Ct) values, with dPCR exhibiting greater sensitivity. We also found that metagenomic sequencing after hybrid selection was best for higher concentration samples while amplicon sequencing performs best for lower concentrations. By establishing these methods, we were able to support the creation of a statewide surveillance program to test bulk milk samples monthly from all cattle dairy farms within Massachusetts, which remain negative to date. The methods, workflow, and recommendations described here provide a framework for others aiming to conduct H5N1 surveillance efforts.
Conflict of interest statement
Conflicts of Interest P.C.S. is co-founder and shareholder in Sherlock Biosciences and Delve Bio, and is a board member and shareholder of Danaher Corporation. DHO is a co-founder and managing member of Pathogenuity LLC.
Figures


References
-
- World Health Organization. Avian Influenza A(H5N1) – United States of America. 2024. April 9 [cited 2024 Oct 1]. https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON512.
-
- US Department of Agriculture Animal and Plant Health Inspection Service. HPAI Confirmed Cases in Livestock. 2024. Jul 3 [cited 2024 Dec 3]. https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influen....
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
