This is a preprint.
Genetic Analysis of Flagellar-Mediated Surface Sensing by Pseudomonas aeruginosa PA14
- PMID: 39677620
- PMCID: PMC11643085
- DOI: 10.1101/2024.12.05.627040
Genetic Analysis of Flagellar-Mediated Surface Sensing by Pseudomonas aeruginosa PA14
Update in
-
Genetic analysis of flagellar-mediated surface sensing by Pseudomonas aeruginosa PA14.J Bacteriol. 2025 Jul 24;207(7):e0052024. doi: 10.1128/jb.00520-24. Epub 2025 Jun 5. J Bacteriol. 2025. PMID: 40470954 Free PMC article.
Abstract
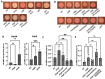
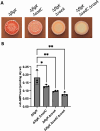
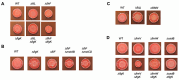
Surface sensing is a key aspect of the early stage of biofilm formation. For P. aeruginosa, the type IV pili (TFP), the TFP alignment complex and PilY1 were shown to play a key role in c-di-GMP signaling upon surface contact. The role of the flagellar machinery in surface sensing is less well understood in P. aeruginosa. Here we show, consistent with findings from other groups, that a mutation in the gene encoding the flagellar hook protein (ΔflgK) or flagellin (ΔfliC) results in a strain that overproduces the Pel exopolysaccharide (EPS) with a concomitant increase in c-di-GMP levels. We use a candidate gene approach and genetic screens, combined with phenotypic assays, to identify key roles for the MotAB and MotCD stators and the FliG protein, a component of the flagellar switch complex, in stimulating the surface-dependent, increased c-di-GMP level noted for these flagellar mutants. These findings are consistent with previous studies showing a role for the stators in surface sensing. We also show that mutations in the genes coding for the diguanylate cyclases SadC and RoeA as well as SadB, a protein involved in early surface colonization, abrogate the increased c-d-GMP-related phenotypes of the ΔflgK mutant. Together, these data indicate that bacteria monitor the status of flagellar synthesis and/or function during surface sensing as a means to trigger the biofilm program.
Keywords: Pseudomonas aeruginosa; biofilm; flagella; stators; surface sensing.
Figures

References
-
- Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
