This is a preprint.
Decoding the biogenesis of HIV-induced CPSF6 puncta and their fusion with the nuclear speckle
- PMID: 39677677
- PMCID: PMC11642789
- DOI: 10.1101/2024.10.06.616889
Decoding the biogenesis of HIV-induced CPSF6 puncta and their fusion with the nuclear speckle
Abstract
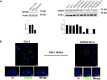
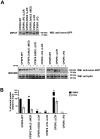
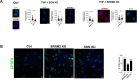
Viruses rely on host cellular machinery for replication. After entering the nucleus, the HIV genome accumulates in nuclear niches where it undergoes reverse transcription and integrates into neighboring chromatin, promoting high transcription rates and new virus progeny. Despite anti-retroviral treatment, viral genomes can persist in these nuclear niches and reactivate if treatment is interrupted, likely contributing to the formation of viral reservoirs. The post-nuclear entry dynamics of HIV remain unclear, and understanding these steps is critical for revealing how viral reservoirs are established. In this study, we elucidate the formation of HIV-induced CPSF6 puncta and the domains of CPSF6 essential for this process. We also explore the roles of nuclear speckle scaffold factors, SON and SRRM2, in the biogenesis of these puncta. Through genetic manipulation and depletion experiments, we demonstrate the key role of the intrinsically disordered region of SRRM2 in enlarging nuclear speckles in the presence of the HIV capsid. We identify the FG domain of CPSF6 as essential for both puncta formation and binding to the viral core, which serves as the scaffold for CPSF6 puncta. While the low-complexity regions (LCRs) modulate CPSF6 binding to the viral capsid, they do not contribute to puncta formation, nor do the disordered mixed charge domains (MCDs) of CPSF6. These results demonstrate how HIV evolved to hijack host nuclear factors, enabling its persistence in the host. Of note, this study provides new insights into the underlying interactions between host factors and viral components, advancing our understanding of HIV nuclear dynamics and offering potential therapeutic targets for preventing viral persistence.
Figures











References
-
- Ay S., and Di Nunzio F. (2023). HIV-Induced CPSF6 Condensates. J Mol Biol 435, 168094. - PubMed
-
- Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., et al. (1983). Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
