This is a preprint.
Convergence of autism proteins at the cilium
- PMID: 39677731
- PMCID: PMC11643032
- DOI: 10.1101/2024.12.05.626924
Convergence of autism proteins at the cilium
Abstract
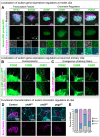
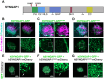
Hundreds of high-confidence autism genes have been identified, yet the relevant etiological mechanisms remain unclear. Gene ontology analyses have repeatedly identified enrichment of proteins with annotated functions in gene expression regulation and neuronal communication. However, proteins are often pleiotropic and these annotations are inherently incomplete. Our recent autism functional genetics work has suggested that these genes may share a common mechanism at the cilium, a membrane-bound organelle critical for neurogenesis, brain patterning, and neuronal activity-all processes strongly implicated in autism. Moreover, autism commonly co-occurs with conditions that are known to involve ciliary-related pathologies, including congenital heart disease, hydrocephalus, and blindness. However, the role of autism genes at the cilium has not been systematically investigated. Here we demonstrate that autism proteins spanning disparate functional annotations converge in expression, localization, and function at cilia, and that patients with pathogenic variants in these genes have cilia-related co-occurring conditions and biomarkers of disrupted ciliary function. This degree of convergence among genes spanning diverse functional annotations strongly suggests that cilia are relevant to autism, as well as to commonly co-occurring conditions, and that this organelle should be explored further for therapeutic potential.
Conflict of interest statement
Competing Interests EB is an employee of Citizen Health with vested and unvested stock options. All other authors do not have any competing interests.
Figures



References
-
- Angerilli Alessandro, Tait Janet, Berges Julian, Shcherbakova Irina, Pokrovsky Daniil, Schauer Tamas, Smialowski Pawel, et al. 2023. “The Histone H4K20 Methyltransferase SUV4–20H1/KMT5B Is Required for Multiciliated Cell Differentiation in Xenopus.” Life Science Alliance 6 (7). 10.26508/lsa.202302023. - DOI - PMC - PubMed
-
- Antoniades Ioanna, Stylianou Panayiota, and Skourides Paris A.. 2014. “Making the Connection: Ciliary Adhesion Complexes Anchor Basal Bodies to the Actin Cytoskeleton.” Developmental Cell 28 (1): 70–80. - PubMed
-
- Bärenz Felix, Mayilo Dmytro, and Gruss Oliver J.. 2011. “Centriolar Satellites: Busy Orbits around the Centrosome.” European Journal of Cell Biology 90 (12): 983–89. - PubMed
-
- Bayés Alex, Collins Mark O., Croning Mike D. R., Lagemaat Louie N. van de, Choudhary Jyoti S., and Grant Seth G. N.. 2012. “Comparative Study of Human and Mouse Postsynaptic Proteomes Finds High Compositional Conservation and Abundance Differences for Key Synaptic Proteins.” PloS One 7 (10): e46683. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
