This is a preprint.
Co-regulator activity of Mediator of DNA Damage Checkpoint 1 (MDC1) is associated with DNA repair dysfunction and PARP inhibitor sensitivity in lobular carcinoma of the breast
- PMID: 39677775
- PMCID: PMC11642799
- DOI: 10.1101/2023.10.29.564555
Co-regulator activity of Mediator of DNA Damage Checkpoint 1 (MDC1) is associated with DNA repair dysfunction and PARP inhibitor sensitivity in lobular carcinoma of the breast
Abstract
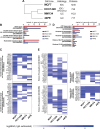
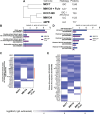
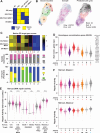
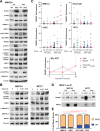
Invasive lobular carcinoma of the breast (ILC) is typically estrogen receptor α (ER)-positive and presents with biomarkers of anti-estrogen sensitive disease, yet patients with ILC face particularly poor long-term outcomes with increased recurrence risk, suggesting endocrine response and ER function are unique in ILC. ER is co-regulated by the DNA repair protein Mediator of DNA Damage Checkpoint 1 (MDC1) specifically in ILC cells, driving distinct ER activity. However, this novel MDC1 activity is associated with dysfunctional canonical DNA repair activity by MDC1, but without typical features of DNA repair deficiency. To understand reciprocal activities of MDC1, we profiled the MDC1 interactome and found MDC1-associated proteins in ILC cells mirror a "BRCA-like" state lacking key homologous recombination (HR) proteins, consistent with HR dysfunction but distinct from classic "BRCAness". HR dysfunction in ILC cells is supported by single-cell transcriptome and DNA repair activity analyses, with DNA repair signaling and functional data, showing dysfunctional induction and resolution of HR. In parallel, ILC tumor data are consistent with a distinct form of HR dysfunction via impaired HR resolution, lacking BRCA-like genomic scarring but showing elevated signatures of PARP inhibitor sensitivity. We demonstrate this HR dysfunction can be exploited using PARP inhibition, and found that talazoparib treatment produced a durable growth suppression both in vitro and in multiple ILC xenografts in vivo. ILC-specific ER:MDC1 activity creates a new context for ER and MDC1 function in ILC, at the cost of a DNA repair dysfunction, which may be therapeutically targetable.
Keywords: DNA damage response; DNA repair; Invasive lobular carcinoma; MDC1; PARP inhibitors; breast cancer; estrogen receptor.
Figures


References
-
- Sikora MJ, Jankowitz RC, Dabbs DJ, Oesterreich S. Invasive lobular carcinoma of the breast: patient response to systemic endocrine therapy and hormone response in model systems. Steroids. 2013. Jun;78(6):568–575. - PubMed
-
- Mouabbi JA, Hassan A, Lim B, Hortobagyi GN, Tripathy D, Layman RM. Invasive lobular carcinoma: an understudied emergent subtype of breast cancer. Breast Cancer Res Treat. 2022. Jun;193(2):253–264. - PubMed
-
- Rakha EA, El-Sayed ME, Powe DG, Green AR, Habashy H, Grainge MJ, et al. Invasive lobular carcinoma of the breast: response to hormonal therapy and outcomes. Eur J Cancer Oxf Engl 1990. 2008. Jan;44(1):73–83. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
