This is a preprint.
Unusually Rapid Isomerization of Aspartic Acid in Tau
- PMID: 39677806
- PMCID: PMC11643016
- DOI: 10.1101/2024.12.04.626870
Unusually Rapid Isomerization of Aspartic Acid in Tau
Update in
-
Determination of Trends Underlying Aspartic Acid Isomerization in Intact Proteins Reveals Unusually Rapid Isomerization of Tau.ACS Chem Neurosci. 2025 Feb 19;16(4):673-686. doi: 10.1021/acschemneuro.4c00721. Epub 2025 Jan 29. ACS Chem Neurosci. 2025. PMID: 39881547 Free PMC article.
Abstract
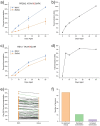
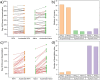
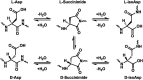
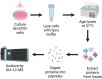
Spontaneous chemical modifications in long-lived proteins can potentially change protein structure in ways that impact proteostasis and cellular health. For example, isomerization of aspartic acid interferes with protein turnover and is anticorrelated with cognitive acuity in Alzheimer's disease. However, few isomerization rates have been determined for Asp residues in intact proteins. To remedy this deficiency, we used protein extracts from SH-SY5Y neuroblastoma cells as a source of a complex, brain-relevant proteome with no baseline isomerization. Cell lysates were aged in vitro to generate isomers, and extracted proteins were analyzed by data-independent acquisition (DIA) liquid chromatography-mass spectrometry (LC-MS). Although no Asp isomers were detected at Day 0, isomerization increased across time and was quantifiable for 105 proteins by Day 50. Data analysis revealed that isomerization rate is influenced by both primary sequence and secondary structure, suggesting that steric hindrance and backbone rigidity modulate isomerization. Additionally, we examined lysates extracted under gentle conditions to preserve protein complexes and found that protein-protein interactions often slow isomerization. Base catalysis was explored as a means to accelerate Asp isomerization due to findings of accelerated asparagine deamidation. However, no substantial rate enhancement was found for isomerization, suggesting fundamental differences in acid-base chemistry. With an enhanced understanding of Asp isomerization in proteins in general, we next sought to better understand Asp isomerization in tau. In vitro aging of monomeric and aggregated recombinant tau revealed that tau isomerizes significantly faster than any similar protein within our dataset, which is likely related to its correlation with cognition in Alzheimer's disease.
Keywords: Alzheimer’s disease; aging; aspartic acid; data-independent acquisition; isomerization; liquid chromatography-mass spectrometry.
Conflict of interest statement
Conflicts of interest. There are no conflicts of interest to declare
Figures






References
-
- Cambridge S. B.; Gnad F.; Nguyen C.; Bermejo J. L.; Krüger M.; Mann M. Systems-Wide Proteomic Analysis in Mammalian Cells Reveals Conserved, Functional Protein Turnover. J. Proteome Res. 2011, 10 (12), 5275–5284. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
