This is a preprint.
Pathogens and Antimicrobial Resistance Genes in Household Environments: A Study of Soil Floors and Cow Dung in Rural Bangladesh
- PMID: 39677809
- PMCID: PMC11642972
- DOI: 10.1101/2024.12.06.627269
Pathogens and Antimicrobial Resistance Genes in Household Environments: A Study of Soil Floors and Cow Dung in Rural Bangladesh
Update in
-
Potential pathogens and antimicrobial resistance genes in household environments: a study of soil floors and cow dung in rural Bangladesh.Appl Environ Microbiol. 2025 Jun 18;91(6):e0066925. doi: 10.1128/aem.00669-25. Epub 2025 May 27. Appl Environ Microbiol. 2025. PMID: 40422288 Free PMC article.
Abstract
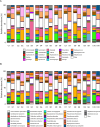
In low- and middle-income countries, living in homes with soil floors and animal cohabitation may expose children to fecal organisms, increasing risk of enteric and antimicrobial-resistant infections. Our objective was to understand whether cow cohabitation in homes with soil floors in rural Bangladesh contributed to the presence and diversity of potential pathogens and antimicrobial resistance genes (ARGs) in the home. In 10 randomly selected households in rural Sirajganj District, we sampled floor soil and cow dung, which is commonly used as sealant in soil floors. We extracted DNA and performed shotgun metagenomic sequencing to explore potential pathogens and ARGs in each sample type. We detected 6 potential pathogens in soil only, 49 pathogens in cow dung only, and 167 pathogens in both soil and cow dung. Pathogen species with relative abundances >5% in both soil floors and cow dung from the same households included E. coli (N=8 households), Salmonella enterica (N=6), Klebsiella pneumoniae (N=2), and Pseudomonas aeruginosa (N=1). Cow dung exhibited modestly higher pathogen genus richness compared to soil floors (Wilcoxon signed-rank test p=0.002). Using Bray-Curtis dissimilarity, pathogen species community composition differed between floors and cow dung (PERMANOVA p<0.001). All soil floors and cow dung samples contained ARGs against antibiotic classes including sulfonamides, rifamycin, aminoglycosides, lincosamides, and tetracycline. Paired floor and cow dung samples shared ARGs against rifamycin. Our findings support the development of interventions to reduce soil and animal feces exposure in rural, low-income settings.
Figures






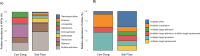
References
-
- Colston JM, Fang B, Nong MK, et al. Spatial variation in housing construction material in low- and middle-income countries: a Bayesian spatial prediction model of a key infectious diseases risk factor and social determinant of health. 2024: 2024.05.23.24307833. - PubMed
-
- Turner R, Hawkes C, Jeff W, et al. Agriculture for improved nutrition: the current research landscape. Food Nutr Bull 2013; 34: 369–77. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
