This is a preprint.
Ancestral Genomic Functional Differences in Oligodendroglia: Implications for Alzheimer's Disease
- PMID: 39678342
- PMCID: PMC11643296
- DOI: 10.21203/rs.3.rs-5338140/v1
Ancestral Genomic Functional Differences in Oligodendroglia: Implications for Alzheimer's Disease
Update in
-
Ancestral genomic functional differences in oligodendroglia: implications for Alzheimer's disease.Alzheimers Dement. 2025 Sep;21(9):e70593. doi: 10.1002/alz.70593. Alzheimers Dement. 2025. PMID: 40937943 Free PMC article.
Abstract
Background: This study aims to elucidate ancestry-specific changes to the genomic regulatory architecture in induced pluripotent stem cell (iPSC)-derived oligodendroglia, focusing on their implications for Alzheimer's disease (AD). This work addresses the lack of diversity in previous iPSC studies by including ancestries that contribute to African American (European/African) and Hispanic/Latino populations (Amerindian/African/European).
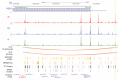
Methods: We generated 12 iPSC lines-four African, four Amerindian, and four European- from both AD patients and non-cognitively impaired individuals, with varying APOE genotypes (APOE3/3 and APOE4/4). These lines were differentiated into neural spheroids containing oligodendrocyte lineage cells. Single-nuclei RNA sequencing and ATAC sequencing were employed to analyze transcriptional and chromatin accessibility profiles, respectively. Differential gene expression, chromatin accessibility, and Hi-C analyses were conducted, followed by pathway analysis to interpret the results.
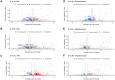
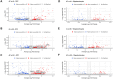
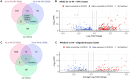
Results: We identified ancestry-specific differences in gene expression and chromatin accessibility. Notably, numerous AD GWAS-associated genes were differentially expressed across ancestries. The largest number of differentially expressed genes (DEGs) were found in European vs. Amerindian and African vs. Amerindian iPSC-derived oligodendrocyte progenitor cells (OPCs). Pathway analysis of APOE4/4 carriers vs APOE3/3 carriers exhibited upregulation of a large number of disease and metabolic pathways in APOE4/4 individuals of all ancestries. Of particular interest was that APOE4/4 carriers had significantly upregulated cholesterol biosynthesis genes relative to APOE3/3 individuals across all ancestries, strongest in iOPCs. Comparison of iOPC and iOL transcriptome data with corresponding human frontal cortex data demonstrated a high correlation (R2 > 0.85).
Conclusions: This research emphasizes the importance of including diverse ancestries in AD research to uncover critical gene expression differences between populations and ancestries that may influence disease susceptibility and therapeutic interventions. The upregulation of cholesterol biosynthesis genes in APOE4/4 carriers of all three ancestries supports the concept that APOE4 may produce disease effects early in life, which could have therapeutic implications as we move forward towards specific therapy for APOE4 carriers. These findings and the high correlation between brain and iPSC-derived OPC and OL transcriptomes support the relevance of this approach as a model for disease study.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures








References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

