This is a preprint.
Identification of specific microbiota members that induce beige fat biogenesis in response to dietary cues
- PMID: 39678347
- PMCID: PMC11643319
- DOI: 10.21203/rs.3.rs-5454144/v1
Identification of specific microbiota members that induce beige fat biogenesis in response to dietary cues
Abstract
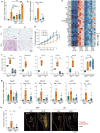
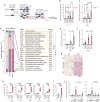
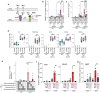
Activation of brown and beige fat biogenesis promotes metabolic health in rodents and humans, but typically requires cold exposure or pharmacological activation of β-adrenergic receptors, which may pose cardiovascular risks. Dietary intervention represents a clinically viable alternative strategy to induce beige cells and thus enhance metabolic health, though the underlying mechanisms remain poorly understood. In this study, we identified specific microbiota members in both mice and humans that promote browning of white adipose tissue (WAT) and ameliorate metabolic disorders in the context of a low-protein diet (LPD). Diets with low protein, regardless of fat and carbohydrate content, induced robust WAT browning to a degree comparable to that achieved by cold exposure or β-adrenergic receptor agonist administration. LPD-mediated browning was markedly diminished in germ-free (GF) mice, highlighting the essential role of the gut microbiota. Microbiota-induced browning occurred independently of the immune system and primarily through mechanisms involving increased circulating deconjugated bile acids, activation of the farnesoid X receptor (FXR) in WAT progenitor cells, and enhanced hepatic production of FGF21. The browning defect in GF mice was rescued by transplanting microbiota from conventional mice or from brown/beige fat-positive healthy human volunteers, as determined by fluorodeoxyglucose positron-emission tomography (FDG-PET). Defined bacterial consortia, comprising strains isolated from mice or FDG-PET-positive human volunteers, were sufficient to elevate plasma bile acids and hepatic FGF21 levels by modulating nitrogen metabolism, ultimately restoring browning in response to a LPD. Our findings highlight the significant impact of diet-microbiota interactions on WAT browning and suggest their therapeutic potential for managing metabolic diseases.
Conflict of interest statement
Additional Declarations: Yes there is potential Competing Interest. Kenya Honda is a scientific advisory board member of Vedanta Biosciences and 4BIO CAPITAL. Masahiro Ueda is an employee of JSR corporation. Ramnik J. Xavier is co-founder of Jnana Therapeutics and Celsius Therapeutics, scientific advisory board member at Nestlé, and board director at MoonLake Immunotherapeutics. Damian.R.Plichta is an employee of Novonesis A/S. All other authors declare no competing interests.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

