This is a preprint.
Multi-dynamic deep image prior for cardiac MRI
- PMID: 39679265
- PMCID: PMC11643223
Multi-dynamic deep image prior for cardiac MRI
Update in
-
Multi-dynamic deep image prior for cardiac MRI.Magn Reson Med. 2025 Dec;94(6):2668-2679. doi: 10.1002/mrm.70000. Epub 2025 Jul 22. Magn Reson Med. 2025. PMID: 40692503 Free PMC article.
Abstract
Purpose: Cardiovascular magnetic resonance imaging is a powerful diagnostic tool for assessing cardiac structure and function. However, traditional breath-held imaging protocols pose challenges for patients with arrhythmias or limited breath-holding capacity. This work aims to overcome these limitations by developing a reconstruction framework that enables high-quality imaging in free-breathing conditions for various dynamic cardiac MRI protocols.
Methods: Multi-Dynamic Deep Image Prior (M-DIP), a novel unsupervised reconstruction framework for accelerated real-time cardiac MRI, is introduced. To capture contrast or content variation, M-DIP first employs a spatial dictionary to synthesize a time-dependent intermediate image. Then, this intermediate image is further refined using time-dependent deformation fields that model cardiac and respiratory motion. Unlike prior DIP-based methods, M-DIP simultaneously captures physiological motion and frame-to-frame content variations, making it applicable to a wide range of dynamic applications.
Results: We validate M-DIP using simulated MRXCAT cine phantom data as well as free-breathing real-time cine, single-shot late gadolinium enhancement (LGE), and first-pass perfusion data from clinical patients. Comparative analyses against state-of-the-art supervised and unsupervised approaches demonstrate M-DIP's performance and versatility. M-DIP achieved better image quality metrics on phantom data, higher reader scores on in-vivo cine and LGE data, and comparable scores on in-vivo perfusion data relative to another DIP-based approach.
Conclusion: M-DIP enables high-quality reconstructions of real-time free-breathing cardiac MRI without requiring external training data. Its ability to model physiological motion and content variations makes it a promising approach for various dynamic imaging applications.
Conflict of interest statement
MV is an employee of Siemens Healthineers AG. FK receives research funding from Siemens Healthineers AG, receives patent royalties for AI for MR image reconstruction from Siemens Healthineers AG, holds stock options from Subtle Medical Inc. and serves as scientific advisor to Imaginostics Inc.
Figures
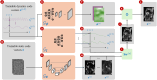
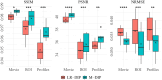

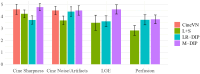

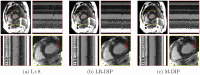


References
-
- Lustig M., Donoho D., Santos J., and Pauly J., “Compressed sensing MRI,” IEEE Signal Process Mag, vol. 25, no. 2, pp. 72–82, 2008, doi: 10.1109/MSP.2007.914728. - DOI
-
- Jalal A., Arvinte M., Daras G., Price E., Dimakis A., and Tamir J., “Robust compressed sensing MRI with deep generative priors,” ser. Proc Neural Inf Process Syst (NeurIPS), vol. 34, Virtual, 2021, pp. 14 938–14 954.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
