The effect of sample site and collection procedure on identification of SARS-CoV-2 infection
- PMID: 39679851
- PMCID: PMC11648846
- DOI: 10.1002/14651858.CD014780
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection
Abstract
Background: Sample collection is a key driver of accuracy in the diagnosis of SARS-CoV-2 infection. Viral load may vary at different anatomical sampling sites and accuracy may be compromised by difficulties obtaining specimens and the expertise of the person taking the sample. It is important to optimise sampling accuracy within cost, safety and accessibility constraints.
Objectives: To compare the sensitivity of different sampling collection sites and methods for the detection of current SARS-CoV-2 infection with any molecular or antigen-based test.
Search methods: Electronic searches of the Cochrane COVID-19 Study Register and the COVID-19 Living Evidence Database from the University of Bern (which includes daily updates from PubMed and Embase and preprints from medRxiv and bioRxiv) were undertaken on 22 February 2022. We included independent evaluations from national reference laboratories, FIND and the Diagnostics Global Health website. We did not apply language restrictions.
Selection criteria: We included studies of symptomatic or asymptomatic people with suspected SARS-CoV-2 infection undergoing testing. We included studies of any design that compared results from different sample types (anatomical location, operator, collection device) collected from the same participant within a 24-hour period.
Data collection and analysis: Within a sample pair, we defined a reference sample and an index sample collected from the same participant within the same clinical encounter (within 24 hours). Where the sample comparison was different anatomical sites, the reference standard was defined as a nasopharyngeal or combined naso/oropharyngeal sample collected into the same sample container and the index sample as the alternative anatomical site. Where the sample comparison was concerned with differences in the sample collection method from the same site, we defined the reference sample as that closest to standard practice for that sample type. Where the sample pair comparison was concerned with differences in personnel collecting the sample, the more skilled or experienced operator was considered the reference sample. Two review authors independently assessed the risk of bias and applicability concerns using the QUADAS-2 and QUADAS-C checklists, tailored to this review. We present estimates of the difference in the sensitivity (reference sample (%) minus index sample sensitivity (%)) in a pair and as an average across studies for each index sampling method using forest plots and tables. We examined heterogeneity between studies according to population (age, symptom status) and index sample (time post-symptom onset, operator expertise, use of transport medium) characteristics.
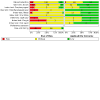
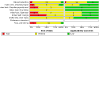
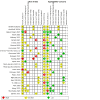
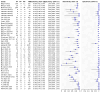
Main results: This review includes 106 studies reporting 154 evaluations and 60,523 sample pair comparisons, of which 11,045 had SARS-CoV-2 infection. Ninety evaluations were of saliva samples, 37 nasal, seven oropharyngeal, six gargle, six oral and four combined nasal/oropharyngeal samples. Four evaluations were of the effect of operator expertise on the accuracy of three different sample types. The majority of included evaluations (146) used molecular tests, of which 140 used RT-PCR (reverse transcription polymerase chain reaction). Eight evaluations were of nasal samples used with Ag-RDTs (rapid antigen tests). The majority of studies were conducted in Europe (35/106, 33%) or the USA (27%) and conducted in dedicated COVID-19 testing clinics or in ambulatory hospital settings (53%). Targeted screening or contact tracing accounted for only 4% of evaluations. Where reported, the majority of evaluations were of adults (91/154, 59%), 28 (18%) were in mixed populations with only seven (4%) in children. The median prevalence of confirmed SARS-CoV-2 was 23% (interquartile (IQR) 13%-40%). Risk of bias and applicability assessment were hampered by poor reporting in 77% and 65% of included studies, respectively. Risk of bias was low across all domains in only 3% of evaluations due to inappropriate inclusion or exclusion criteria, unclear recruitment, lack of blinding, nonrandomised sampling order or differences in testing kit within a sample pair. Sixty-eight percent of evaluation cohorts were judged as being at high or unclear applicability concern either due to inflation of the prevalence of SARS-CoV-2 infection in study populations by selectively including individuals with confirmed PCR-positive samples or because there was insufficient detail to allow replication of sample collection. When used with RT-PCR • There was no evidence of a difference in sensitivity between gargle and nasopharyngeal samples (on average -1 percentage points, 95% CI -5 to +2, based on 6 evaluations, 2138 sample pairs, of which 389 had SARS-CoV-2). • There was no evidence of a difference in sensitivity between saliva collection from the deep throat and nasopharyngeal samples (on average +10 percentage points, 95% CI -1 to +21, based on 2192 sample pairs, of which 730 had SARS-CoV-2). • There was evidence that saliva collection using spitting, drooling or salivating was on average -12 percentage points less sensitive (95% CI -16 to -8, based on 27,253 sample pairs, of which 4636 had SARS-CoV-2) compared to nasopharyngeal samples. We did not find any evidence of a difference in the sensitivity of saliva collected using spitting, drooling or salivating (sensitivity difference: range from -13 percentage points (spit) to -21 percentage points (salivate)). • Nasal samples (anterior and mid-turbinate collection combined) were, on average, 12 percentage points less sensitive compared to nasopharyngeal samples (95% CI -17 to -7), based on 9291 sample pairs, of which 1485 had SARS-CoV-2. We did not find any evidence of a difference in sensitivity between nasal samples collected from the mid-turbinates (3942 sample pairs) or from the anterior nares (8272 sample pairs). • There was evidence that oropharyngeal samples were, on average, 17 percentage points less sensitive than nasopharyngeal samples (95% CI -29 to -5), based on seven evaluations, 2522 sample pairs, of which 511 had SARS-CoV-2. A much smaller volume of evidence was available for combined nasal/oropharyngeal samples and oral samples. Age, symptom status and use of transport media do not appear to affect the sensitivity of saliva samples and nasal samples. When used with Ag-RDTs • There was no evidence of a difference in sensitivity between nasal samples compared to nasopharyngeal samples (sensitivity, on average, 0 percentage points -0.2 to +0.2, based on 3688 sample pairs, of which 535 had SARS-CoV-2).
Authors' conclusions: When used with RT-PCR, there is no evidence for a difference in sensitivity of self-collected gargle or deep-throat saliva samples compared to nasopharyngeal samples collected by healthcare workers when used with RT-PCR. Use of these alternative, self-collected sample types has the potential to reduce cost and discomfort and improve the safety of sampling by reducing risk of transmission from aerosol spread which occurs as a result of coughing and gagging during the nasopharyngeal or oropharyngeal sample collection procedure. This may, in turn, improve access to and uptake of testing. Other types of saliva, nasal, oral and oropharyngeal samples are, on average, less sensitive compared to healthcare worker-collected nasopharyngeal samples, and it is unlikely that sensitivities of this magnitude would be acceptable for confirmation of SARS-CoV-2 infection with RT-PCR. When used with Ag-RDTs, there is no evidence of a difference in sensitivity between nasal samples and healthcare worker-collected nasopharyngeal samples for detecting SARS-CoV-2. The implications of this for self-testing are unclear as evaluations did not report whether nasal samples were self-collected or collected by healthcare workers. Further research is needed in asymptomatic individuals, children and in Ag-RDTs, and to investigate the effect of operator expertise on accuracy. Quality assessment of the evidence base underpinning these conclusions was restricted by poor reporting. There is a need for further high-quality studies, adhering to reporting standards for test accuracy studies.
Copyright © 2024 The Authors. Cochrane Database of Systematic Reviews published by John Wiley & Sons, Ltd. on behalf of The Cochrane Collaboration.
Conflict of interest statement
Jonathan J Deeks*: none known.
Jacqueline Dinnes (JD)* is a Cochrane DTA Editor. She was not involved in the editorial process for this review. JD declares a grant from Evidence Synthesis Ireland, National University of Ireland, Galway, to supervise three fellows contributing to a Cochrane review of rapid antigen tests for Covid‐19; paid to the institution. JD declares publishing opinions on the topic: (1) Dinnes J, Davenport C. COVID‐19 rapid antigen testing strategies must be evaluated in intended use settings. Lancet. Regional Health, Western Pacific 2022;25:100542. DOI: 10.1016/j.lanwpc.2022.100542; (2) Deeks JJ, Dinnes J, Davenport C, Takwoingi Y, McInnes M, Leeflang MM, Cunningham J. Letter to the Editor regarding Peto T; UK COVID‐19 Lateral Flow Oversight Team: COVID‐19: Rapid antigen detection for SARS‐CoV‐2 by lateral flow assay. EClinicalMedicine 2021;38; (3) Dinnes J. COVID‐19 rapid antigen testing strategies require careful evaluation. EBioMedicine 2021;70:103491; and (4) Dinnes J, Davenport C. Do we have informed consent for asymptomatic COVID‐19 testing in schools? BMJ Opinion. Available at https://blogs.bmj.com/bmj/2021/03/16/do‐we‐have‐informed‐consent‐for‐asymptomatic‐testing‐in‐schools/, 2021 [https://blogs.bmj.com/bmj/2021/03/16/do‐we‐have‐informed‐consent‐for‐asymptomatic‐testing‐in‐schools/, 2021].
Yemisi Takwoingi* is a member of the Cochrane Editorial Board, and an Editor with Cochrane Infectious Diseases and the Cochrane DTA Editorial Team. She was not involved in the editorial process for this review.
Clare Davenport* is the Contact Editor for the Cochrane DTA Editorial Team. She was not involved in the editorial process for this review.
Mariska MG Leeflang (MML) is an Associate Professor at Academisch Medisch Centrum, and Editor and member of the DTA Editorial Team. She was not involved in the editorial process for this review. MMGL declares a grant from Cochrane for finalising the DTA Handbook, of which she is an active proponent, and royalties from the sale of the Handbook; personal payment.
René Spijker (RS) is employed as an Information Specialist for three days per week at Amsterdam UMC. For two days per week, she is seconded to Cochrane Netherlands, which is hosted by the Universitair Medisch Centrum Utrecht. She was not involved in the editorial process for this review. RS declares that the Dutch Cochrane Centre (DCC) has received grants for performing commissioned systematic reviews. In no situation, the commissioner had any influence on the results of the work.
Sarah Berhane* is employed as a medicial statistician via grant funding from NIHR Birmingham Biomedical Research Centre to the University of Birmingham.
Ann Van den Bruel: none known.
Devy Emperador: is employed by FIND, with funding from FCDO and CAN. FIND is a global non‐for‐profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high‐quality diagnostic tools for low‐resource settings and this is achieved by supporting both research and development and access activities for a wide range of diseases, including COVID‐19. FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review (i.e. these companies donate these tests to FIND for free for Find‐sponsored research studies).
Ingrid Arevalo‐Rodriguez: none known.
Miriam Mateos‐Haro: none known.
Agustin Ciapponi works as a health professional at Servicio de Medicina Familiar y Comunitaria, Hospital Italiano de Buenos Aires, Argentina.
Diana Buitrago‐Garcia: none known.
*Jonathan Deeks, Jacqueline Dinnes, Yemisi Takwoingi, Clare Davenport and Sarah Berhane are supported by the NIHR Birmingham Biomedical Research Centre. This paper presents independent research supported by the NIHR Birmingham Biomedical Research Centre at the University Hospitals Birmingham NHS Foundation Trust and the University of Birmingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Figures




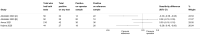
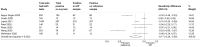















Similar articles
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Thoracic imaging tests for the diagnosis of COVID-19.Cochrane Database Syst Rev. 2022 May 16;5(5):CD013639. doi: 10.1002/14651858.CD013639.pub5. Cochrane Database Syst Rev. 2022. PMID: 35575286 Free PMC article.
Cited by
-
A scoping review on the body awareness rehabilitation after stroke: are we aware of what we are unaware?Front Neurol. 2025 Jan 7;15:1497052. doi: 10.3389/fneur.2024.1497052. eCollection 2024. Front Neurol. 2025. PMID: 39839868 Free PMC article. Review.
References
References to studies included in this review
Abdollahi 2020a {published data only}
Abdollahi 2020b {published data only}
AbdulRahman 2020a {published data only}
-
- AbdulRahman AK, AlBastaki A, AlAwadhi A, Alwazaan A, AlQahtani M. Diagnostic and monitoring utilities of saliva for SARS-CoV-2. MedRxiv 2020;NA:no pagination. [DOI: ]
AbdulRahman 2020b {published data only}
-
- AbdulRahman A, AlBastaki A, AlAwadhi A, Alwazaan A, AlQahtani M. Diagnostic and monitoring utilities of saliva for SARS-CoV-2. MedRxiv 2020;NA:no pagination. [DOI: ]
Aita 2020 {published data only}
Akgun Dogan 2021 {published data only}
Al Suwaidi 2021 {published data only}
Altawalah 2020 {published data only}
Amoo 2021 {published data only}
Babady 2021 {published data only}
Banerjee 2021 {published data only}
Becker 2020 {published data only}
-
- Becker D, Sandoval E, Amin A, De Hoff P, Diets A, Leonetti N, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. MedRxiv 2020;NA:no pagination. [DOI: ]
Begum 2022 {published data only}
Benoit 2021 {published data only}
-
- Benoit P, Labbe AC, Lalancette L, Gagnon S, Bonneau E, Lavallee C, et al. Comparison of SARS-CoV-2 detection with the Cobas(R) 6800/8800 system on gargle samples using two sample processing methods with combined oropharyngeal/nasopharyngeal swab. Journal of Medical Virology 2021;93(12):6837-40. - PMC - PubMed
Bhattacharya 2021 {published data only}
-
- Bhattacharya D, Parai D, Rout UK, Dash P, Nanda RR, Dash GC, et al. Saliva for diagnosis of SARS-CoV-2: first report from India. Journal of Medical Virology 2021;93(4):2529-33. - PubMed
Bidkar 2022 {published data only}
Bikos 2021 {published data only}
-
- Bikos DA, Hwang C, Brileya KA, Parker A, Loveday EK, Rodriguez M, et al. SLAMP: a rapid fluorometric RT-LAMP assay for sensitive and specific detection of SARS-CoV-2 from human saliva. MedRxiv 2021;NA:no pagination. [DOI: ]
Boerger 2021 {published data only}
Borghi 2021a {published data only}
Borghi 2021b {published data only}
Borghi 2021c {published data only}
Boutros 2021(a) {published data only}
Boutros 2021 (b) {published data only}
Bruxvoort 2021 {published data only}
Callahan 2021(a) {published data only}
Callahan 2021(b) {published data only}
Callahan 2021(c) {published data only}
Callahan 2021(d) {published data only}
Callahan 2021(e) {published data only}
Castelain 2021 {published data only}
-
- Castelain S, Francois C, Demey B, Aubry A, Lanoix JP, Duverlie G, et al. Comparison of nasopharyngeal and saliva swabs for the detection of RNA SARS-CoV-2 during mass screening (SALICOV study). New Microbiologica 2021;44(1):59-61. - PubMed
Chu 2021 (a) {published data only}
Chu 2021 (b) {published data only}
Congrave‐Wilson 2021 {published data only}
Cradic 2020 {published data only}
Da Costa Fernandes 2021 {published data only}
Da Silva 2022 {published data only}
-
- Verissimo Rodrigues Da Silva N, Neves Pereira SS, Kavalco KF, Bezerra Meneg�dio F, Medeiros-Santana L, Vis�tto LE, et al. Molecular diagnosis of SARS-CoV-2: a validation of saliva samples. MedRxiv 2022;NA:no pagination. [DOI: ]
De Oliveira 2021 {published data only}
Desai 2021(a)[A] {published data only}
Desai 2021(a)[B] {published data only}
Desai 2021(a)[C] {published data only}
Desai 2021(b) {published data only}
Desmet 2020 {published data only}
Dumaresq 2021 {published data only}
Echavarria 2021 {published data only}
Escobar 2021 {published data only}
Felix 2022 {published data only}
-
- Felix AC, De Paula AV, Ribeiro AC, Da Silva FC, Inemami M, Costa AA, et al. Saliva as a reliable sample for COVID-19 diagnosis in paediatric patients. International Journal of Paediatric Dentistry 2022;32(1):123-5. - PubMed
Fernandez‐Gonzalez 2021(a) {published data only}
Fernandez‐Gonzalez 2021(b) {published data only}
Fernandez‐Gonzalez 2021 (c) {published data only}
Fernández‐Pittol 2020 {published data only}
-
- Fernandez-Pittol M, Hurtado JC, Moreno-Garcia E, Rubio E, Navarro M, Valiente M, et al. Assessment of the use and quick preparation of saliva for rapid microbiological diagnosis of COVID-19. BioRxiv 2020;NA:no pagination. [DOI: ]
FIND (a) 2021 {published data only}
-
- FIND. Evaluation of bionote, inc. Nowcheck COVID-19 Ag test nasal. External report. https://www.finddx.org/wp-content/uploads/2023/01/20220923_bionote_nowch... (accessed prior to 26/10/2024);(1.0).
FIND (b) 2021 {published data only}
-
- FIND. Evaluation of Abbott Panbio COVID-19ag rapid test device (nasal). https://pmc.ncbi.nlm.nih.gov/articles/PMC8111145/ (accessed prior to 26/10/2024).
FIND (c) 2021(a) {published data only}
-
- FIND. Evaluation of SD biosensor, inc. STANDARD q COVID-19 ag test, nasal swab. External report. https://www.finddx.org/wp-content/uploads/2023/01/20221004_sd_biosensor_... 2021;(1.0).
FIND(c) 2021 (b) {published data only}
-
- Evaluation of SD biosensor, inc. STANDARD q COVID-19 ag test, nasal swab. https://www.finddx.org/wp-content/uploads/2023/01/20210413_sd_biosensor_... (accessed prior to 26/10/2024);(1.0).
FIND(s) 2021 {published data only}
-
- FIND. Evaluation of Premier Medical Corporation Pvt.Ltd Sure Status COVID-19 Antigen Card test, nasal swab. External report. https://www.finddx.org/wp-content/uploads/2023/01/20211201_premier_sures... (accessed prior to 26/10/2024);(1.0).
Fougere 2021 {published data only}
-
- Fougere Y, Schwob JM, Miauton A, Hoegger F, Opota O, Jaton K, et al. Performance of RT-PCR on saliva specimens compared with nasopharyngeal swabs for the detection of SARS-CoV-2 in children: a prospective comparative clinical trial. Pediatric Infectious Disease Journal 2021;40(8):e300-4. - PubMed
Fronza 2021 (a) {published data only}
Fronza 2021 (b) {published data only}
Goncalves 2021 {published data only}
Griesemer 2021 (a) {published data only}
Griesemer 2021 (b) {published data only}
Guclu 2020 (a) {published data only}
-
- Guclu E, Koroglu M, Yurumez Y, Toptan H, Kose E, Guneysu F, et al. Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Revista da Associação Médica Brasileira 2020;66(8):1116-21. - PubMed
Guclu 2020 (b) {published data only}
-
- Guclu E, Koroglu M, Yurumez Y, Toptan H, Kose E, Guneysu F, et al. Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Revista da Associação Médica Brasileira 2020;66(8):1116-21. - PubMed
Guzman‐Ortiz 2021 {published data only}
-
- Guzman-Ortiz AL, Nevfrez-Ramirez AJ, Lopez-Martinez B, Parra-Ortega I, Angeles-Floriano T, Martinez-Rodriguez N, et al. Sensitivity of the molecular test in saliva for detectionof COVID-19 in pediatric patients with concurrent conditions. Frontiers in Paediatrics 2021;9:642781. [DOI: 10.3389/fped.2021.642781] - DOI - PMC - PubMed
Hamilton 2021 {published data only}
Herrera 2021 {published data only}
Huber 2021 (a) {published data only}
Huber 2021 (b) {published data only}
Huber 2021 (c) {published data only}
Justo 2021 {published data only}
Kannian 2021 {published data only}
Kerneis 2021 (a) {published data only}
-
- Kerneis S, Elie C, Fourgeaud J, Choupeaux L, Delarue SM, Alby ML, et al. Accuracy of saliva and nasopharyngeal sampling for detection of SARS-CoV-2 in community screening: a multicentric cohort study. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(11):2379-88. - PMC - PubMed
Kerneis 2021 (b) {published data only}
-
- Kerneis S, Elie C, Fourgeaud J, Choupeaux L, Delarue SM, Alby ML, et al. Accuracy of saliva and nasopharyngeal sampling for detection of SARS-CoV-2 in community screening: a multicentric cohort study. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(11):2379-88. - PMC - PubMed
Kerneis 2021 (c) {published data only}
-
- Kerneis S, Elie C, Fourgeaud J, Choupeaux L, Delarue SM, Alby ML, et al. Accuracy of saliva and nasopharyngeal sampling for detection of SARS-CoV-2 in community screening: a multicentric cohort study. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(11):2379-88. - PMC - PubMed
Klein 2021 {published data only}
-
- Klein JA, Kruger LJ, Tobian F, Gaeddert M, Lainati F, Schnitzler P, et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Medical Microbiology and Immunology 2021;210(4):181-6. - PMC - PubMed
Kocagoz 2021 {published data only}
-
- Kocagoz T, Can O, Yurttutan Uyar N, Aksoy E, Polat T, Cankaya D, et al. Simple concentration method enables the use of gargle and mouthwash instead of nasopharyngeal swab sampling for the diagnosis of COVID-19 by PCR. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(12):2617-22. - PMC - PubMed
Kojima 2020 {published data only}
Labhardt 2022 {published data only}
Landry 2020 {published data only}
Leung 2020 {published data only}
Lindner 2021 {published data only}
Lopes 2021 {published data only}
Mahmoud 2021 {published data only}
Marquette 2021 (a) {published data only}
Marquette 2021 (b) {published data only}
Marquette 2021 (c) {published data only}
Marx 2021 (a) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (b) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (c) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (d) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (e) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (f) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (g) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (h) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (i) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (j) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (k) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (l) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (m) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (n) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (o) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (p) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (q) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Marx 2021 (r) {published data only}
-
- Marx GE, Smith-Jeffcoat SE, Biggerstaff BJ, Koh M, Nawrocki CC, Travanty EA, et al. SARS-CoV-2 detection by rRT-PCR on self-collected anterior nares swabs or saliva compared with clinician-collected nasopharyngeal swabs - Denver and Atlanta, August-November, 2020. MedRxiv 2021;NA:no pagination. [DOI: ]
Matic 2021 {published data only}
McCormick‐Baw 2020 {published data only}
McCulloch 2020 {published data only}
McLennan 2022 {published data only}
Melo Costa 2021 (a) {published data only}
Melo Costa 2021 (b) {published data only}
Melo Costa 2021 (c) {published data only}
Mestdagh 2021 (a)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (a)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (b)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (b)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (c)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (c)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (d)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (d)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (e)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (e)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (f)[A] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Mestdagh 2021 (f)[B] {published data only}
-
- Mestdagh P, Gillard M, Dhillon SK, Pirnay JP, Poels J, Hellemans J, et al. Evaluating diagnostic accuracy of saliva sampling methods for severe acute respiratory syndrome coronavirus 2 reveals differential sensitivity and association with viral load. Journal of Molecular Diagnostics 2021;23(10):1249-58. - PMC - PubMed
Migueres 2020 {published data only}
Migueres 2021 (a) {published data only}
Migueres 2021 (b) {published data only}
Migueres 2021 (c) {published data only}
Migueres 2021 (d) {published data only}
Migueres 2021 (e) {published data only}
Migueres 2021 (f) {published data only}
Miller 2020 {published data only}
-
- Miller M, Jansen M, Bisignano A, Mahoney S, Wechsberg C, Albanese N, et al. Validation of a self-administrable, saliva-based RT-qpcr test detecting SARS-CoV-2. MedRxiv 2020;NA:no pagination. [DOI: ]
Mora‐Aguilera 2022 (a) {published data only}
-
- Mora-Aguilera G, Martinez-Bustamante V, Acevedo-Sanchez G, Coria-Contreras JJ, Guzman-Hernandez E, Flores-Colorado OE, et al. Surveillance web system and mouthwash-saliva qPCR for labor ambulatory SARS-CoV-2 detection and prevention. International Journal of Environmental Research and Public Health 2022;19(3):551-7. - PMC - PubMed
Mora‐Aguilera 2022 (b) {published data only}
-
- Mora-Aguilera G, Martinez-Bustamante V, Acevedo-Sanchez G, Coria-Contreras JJ, Guzman-Hernandez E, Flores-Colorado OE, et al. Surveillance web system and mouthwash-saliva qPCR for labor ambulatory SARS-CoV-2 detection and prevention. International Journal of Environmental Research and Public Health 2022;19(3):551-7. - PMC - PubMed
Moreno‐Contreras 2020 {published data only}
-
- Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, Cantu-Cuevas MA, Baron-Olivares H, Ortiz-Orozco OD, et al. Saliva sampling and its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages. Journal of Clinical Microbiology 2020;58(10):e01659-20. - PMC - PubMed
Nacher 2021 (a) {published data only}
-
- Nacher M, Mergeay-Fabre M, Blanchet D, Benois O, Pozl T, Mesphoule P, et al. Diagnostic accuracy and acceptability of molecular diagnosis of COVID-19 on saliva samples relative to nasopharyngeal swabs in tropical hospital and extra-hospital contexts: the COVISAL study. PloS One 2021;16(9):e0257169. - PMC - PubMed
Nacher 2021 (b) {published data only}
-
- Nacher M, Mergeay-Fabre M, Blanchet D, Benois O, Pozl T, Mesphoule P, et al. Diagnostic accuracy and acceptability of molecular diagnosis of COVID-19 on saliva samples relative to nasopharyngeal swabs in tropical hospital and extra-hospital contexts: the COVISAL study. PloS One 2021;16(9):e0257169. - PMC - PubMed
Otto 2020 {published data only}
Pasomsub 2020 {published data only}
Patel 2020 {published data only}
Patriquin 2022 {published data only}
-
- Patriquin G, LeBlanc JJ, Gillis HA, McCracken GR, Pettipas JJ, Hatchette TF. Combined oropharyngeal/nares and nasopharyngeal swab sampling remain effective for molecular detection of SARS-CoV-2 Omicron variant. Journal of Medical Microbiology 2022;71(6):288. - PubMed
Pere 2020 {published data only}
Pham 2020 {published data only}
Potter 2022 {published data only}
-
- Potter RF, Ransom EM, Wallace MA, Johnson C, Kwon JH, Babcock HM, et al. Multiplatform assessment of saliva for SARS-CoV-2 molecular detection in symptomatic healthcare personnel and patients presenting to the emergency department. Journal of Applied Laboratory Medicine 2022;7(3):727-36. - PMC - PubMed
Procop 2020 {published data only}
Rajh 2021[A] {published data only}
Rajh 2021[B] {published data only}
Rajh 2021[C] {published data only}
Rao 2021(a) {published data only}
Rao 2021 (b) {published data only}
Sahajpal 2021 {published data only}
-
- Sahajpal NS, Mondal AK, Ananth S, Njau A, Ahluwalia P, Kota V, et al. SalivaAll: clinical validation of a sensitive test for saliva collected in healthcare and community settings with pooling utility for SARS-CoV-2 mass surveillance. Journal of Molecular Diagnostics 2021;23(7):788-95. - PMC - PubMed
Sahni 2021 {published data only}
-
- Sahni LC, Avadhanula V, Ortiz CS, Feliz KE, John RE, Brown CA, et al. Comparison of mid-turbinate and nasopharyngeal specimens for molecular detection of SARS-CoV-2 among symptomatic outpatients at a pediatric drive-through testing site. Journal of the Pediatric Infectious Diseases Society 2021;10(8):872-9. - PubMed
Salvatore 2021 {published data only}
Schwob 2020 {published data only}
Senok 2020 {published data only}
Skolimowska 2020 {published data only}
Smith‐Jeffcoat 2021 {published data only}
Teo 2021 (a) {published data only}
Teo 2021 (b) {published data only}
Torres 2021 {published data only}
Tu 2020 [A] {published data only}
-
- Tu YP, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, et al. Patient-collected tongue, nasal, and mid-turbinate swabs for SARS-CoV-2 yield equivalent sensitivity to health care worker collected nasopharyngeal swabs. MedRxiv 2020;NA:no pagination. [DOI: ]
Tu 2020 [B] {published data only}
Tu 2020 [C] {published data only}
-
- Tu YP, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, et al. Patient-collected tongue, nasal, and mid-turbinate swabs for SARS-CoV-2 yield equivalent sensitivity to health care worker collected nasopharyngeal swabs. MedRxiv [Preprint] 2020;NA:1-20.
Uddin 2021 {published data only}
Vaz 2020 {published data only}
Vermeiren 2020 {published data only}
Villar 2020 {published data only}
Vlek 2020 {published data only}
Walker 2021 {published data only}
Wang 2020 {published data only}
Wehrhahn 2020 {published data only}
Williams 2020 {published data only}
References to studies excluded from this review
Alghounaim 2020 {published data only}
-
- Alghounaim M, Almazeedi S, Al Youha S, Papenburg J, Alowaish O, AbdulHussain G, et al. Low-cost polyester-tipped three-dimensionally printed nasopharyngeal swab for the detection of severe acute respiratory syndrome-related coronavirus 2 (SARS-cov-2). Journal of Clinical Microbiology 2020;58(11):e01668-20. - PMC - PubMed
Altamirano 2020 {published data only}
Avaniss‐Aghajani 2020 {published data only}
-
- Avaniss-Aghajani E, Sarkissian A, Fernando F, Avaniss-Aghajani A. Validation of the hologic Aptima unisex and multitest specimen collection kits used for endocervical and male urethral swab specimens (Aptima swabs) for collection of samples from SARS-CoV-2-infected patients. Journal of Clinical Microbiology 2020;58(8):e00753-20. - PMC - PubMed
Azzi 2020 {published data only}
Basso 2020 {published data only}
-
- Basso D, Aita A, Navaglia F, Franchin E, Fioretto P, Moz S, et al. SARS-CoV-2 RNA identification in nasopharyngeal swabs: issues in pre-analytics. Clinical Chemistry and Laboratory Medicine 2020;58(9):1579-86. - PubMed
Begum 2022a {published data only}
Bennett 2020 {published data only}
-
- Bennett I, Bulterys PL, Chang M, DeSimone JM, Fralick J, Herring M, et al. The rapid deployment of a 3D printed latticed nasopharyngeal swab for COVID-19 testing made using digital light synthesis. MedRxiv 2020;NA:no pagination. [DOI: 10.1101/2020.05.25.20112201] - DOI
Berenger 2020 {published data only}
-
- Berenger BM, Fonseca K, Schneider AR, Hu J, Zelyas N. Sensitivity of nasopharyngeal, nasal and throat swab for the detection of SARS-CoV-2. MedRxiv 2020;NA:1-10.
Berenger 2021 {published data only}
Bergevin 2021 {published data only}
-
- Bergevin MA, Freppel W, Robert G, Ambaraghassi G, Aubry D, Haeck O, et al. Validation of saliva sampling as an alternative to oro-nasopharyngeal swab for detection of SARS-CoV-2 using unextracted rRT-PCR with the Allplex 2019-nCoV assay. Journal of Medical Microbiology 2021;70(8):001404. - PMC - PubMed
Beyene 2021 {published data only}
Bland 2021 {published data only}
Bloom 2021 {published data only}
-
- Bloom JS, Sathe L, Munugala C, Jones EM, Gasperini M, Lubock NB, et al. Swab-Seq: a high-throughput platform for massively scaled up SARS-CoV-2 testing. MedRxiv 2021;NA:no pagination. [DOI: ]
Braz‐Silva 2020 {published data only}
Brotons 2020 {published data only}
Bulfoni 2021 {published data only}
Bulfoni 2022 {published data only}
Bundgaard 2021 {published data only}
Byrne 2020 {published data only}
Calame 2020 {published data only}
Callahan 2020 {published data only}
-
- Callahan C, Lee R, Lee G, Zulauf KE, Kirby JE, Arnaout R. Nasal-swab testing misses patients with low SARS-CoV-2 viral loads. MedRxiv 2020;NA:1-19.
Carmagnola 2021 {published data only}
Carvalho 2021 {published data only}
Casati 2021 {published data only}
-
- Casati B, Verdi JP, Hempelmann A, Kittel M, Klaebisch AG, Meister B, et al. ADESSO detects SARS-CoV-2 and its variants: extensive clinical validation of an optimised CRISPR-Cas13-based COVID-19 test. MedRxiv 2021;NA:no pagination. [DOI: ]
Cassinari 2021 {published data only}
Caulley 2020 {published data only}
Chan 2020a {published data only}
-
- Chan JF, Yip CC, To KK, Tang TH, Wong SC, Leung KH, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. Journal of Clinical Microbiology 2020;58(5):e00310-20. - PMC - PubMed
Chan 2020b {published data only}
Chapleau 2020 {published data only}
-
- Chapleau RR, Fries AC, Lisanby MW, Rhode MG, Salisbury R, Starr CR. Alternatives to viral transport medium for use in SARS-cov-2 sample preparation. Journal of Clinical and Diagnostic Research 2020;14(12):Lc07-10.
Chen 2020 {published data only}
Cheuk 2020 {published data only}
Chivte 2021 {published data only}
Chong 2020 {published data only}
Chotimah 2022 {published data only}
-
- Chotimah SN, Putra YE, JH Ng, Tijptaningrum A, Sahara NS, Arfianti E, et al. DNA Aptamer gold nanoparticle colorimetric diagnostic test kit of saliva samples for SARS-CoV-2 virus linked to mobile phone application (Aptamextm). MedRxiv 2022;NA:no pagination. [DOI: ]
Chu 2021 {published data only}
Clementino 2022 {published data only}
-
- Clementino M, Cavalcante KF, Viana VAF, Silva DO, Damasceno CR, Fernandes de Souza J, et al. Detection of SARS-CoV-2 in different human biofluids using the loop-mediated isothermal amplification assay: a prospective diagnostic study in Fortaleza, Brazil. Journal of Medical Virology 2022;94(9):4170-80. - PMC - PubMed
Comerlato 2022 {published data only}
Connor 2022 {published data only}
Costa 2021 {published data only}
Costeloe 2021 {published data only}
-
- Costeloe A, Samad MN, Babu S, Metz C. Comparison of tracheal vs nasopharyngeal secretions for SARS-CoV-2 RT-PCR testing in patients with tracheostomy. Otolaryngology and Head and Neck Surgery 2021;165(1):89-92. - PubMed
D'Andrea 2021 {published data only}
De Ioris 2020 {published data only}
-
- De Ioris MA, Scarselli A, Ciofi Degli Atti ML, Rava L, Smarrazzo A, Concato C, et al. Dynamic viral severe acute respiratory syndrome Coronavirus-2 RNA shedding in children: preliminary data and clinical consideration from a Italian regional center. Journal of the Pediatric Infectious Diseases Society 2020;9(3):366-9. - PMC - PubMed
Delaney 2020 {published data only}
-
- Delaney M, Simpson J, Thomas B, Ralph C, Evangalista M, Moshgriz M, et al. The use of saliva as a diagnostic specimen for SARS CoV-2 molecular diagnostic testing for pediatric patients. MedRxiv 2020;NA:no pagination. [DOI: ]
De Marinis 2021 {published data only}
De Marinis 2021a {published data only}
De Oliveira 2022 {published data only}
De Santi 2021 {published data only}
Dhakad 2021 {published data only}
Di Pietro 2020 {published data only}
-
- Capecchi E, Di Pietro GM, Luconi E, Testing Pediatric COVID-19. Is nasopharyngeal swab comparable with nasopharyngeal aspirate to detect SARS-CoV-2 in children? Pediatric Infectious Disease Journal 2020;39(9):e288-9. - PubMed
Donato 2021 {published data only}
-
- Donato LJ, Trivedi VA, Stransky AM, Misra A, Pritt BS, Binnicker MJ, et al. Evaluation of the Cue Health point-of-care COVID-19 (SARS-CoV-2 nucleic acid amplification) test at a community drive-through collection center. Diagnostic Microbiology and Infectious Disease 2021;100(1):115307. - PMC - PubMed
Esteves 2022 {published data only}
Fabiani 2022 {published data only}
-
- Fabiani L, Mazzaracchio V, Moscone D, Fillo S, De Santis R, Monte A, et al. Paper-based immunoassay based on 96-well wax-printed paper plate combined with magnetic beads and colorimetric smartphone-assisted measure for reliable detection of SARS-CoV-2 in saliva. Biosensors & Bioelectronics 2022;200:113909. - PMC - PubMed
Fan 2020 {published data only}
Fang 2020 {published data only}
Federman 2020 {published data only}
Fougere 2021a {published data only}
-
- Fougere Y, Schwob JM, Miauton A, Hoegger F, Opota O, Jaton K, et al. Performance of RT-PCR on saliva specimens compared with nasopharyngeal swabs for the detection of SARS-CoV-2 in children: a prospective comparative clinical trial. Pediatric Infectious Disease Journal 2021;40(8):e300-4. - PubMed
Fowler 2020 {published data only}
-
- Fowler VL, Armson B, Gonzales JL, Wise EL, Howson EL, Vincent-Mistiaen Z, et al. A reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 within nasopharyngeal and oropharyngeal swabs at Hampshire Hospitals NHS Foundation Trust. MedRxiv 2020;NA:no pagination. [DOI: ]
Freire‐Paspuel 2020 {published data only}
Fronza 2022 {published data only}
Gable 2021 {published data only}
-
- Gable P, Huang JY, Gilbert SE, Bollinger S, Lyons AK, Sabour S, et al. A comparison of less invasive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostic specimens in nursing home residents - Arkansas, June-August 2020. Clinical Infectious Diseases 2021;73(Suppl 1):S58-64. - PMC - PubMed
Gan 2020 {published data only}
-
- Gan X, Hua L, Liu Q, Xie D, Wu Z, Xiong Y, et al. Clinical value of anal swab positive in COVID-19 patients. Chinese Journal of Microbiology and Immunolology 2020;40(7):489-94.
Georgas 2022 {published data only}
Girish 2021 {published data only}
-
- Girish P, Jayasankar P, Abhishek P, Sumeeta S, Gunvant P, Shalin P. Comparative analysis of the naso/oropharyngeal swab and oral bio-fluid (whole saliva) samples for the detection of SARS-CoV-2 using RT-qPCR. Indian Journal of Dental Research 2021;32(2):206-10. - PubMed
Goldfarb 2020 {published data only}
Goodall 2022 {published data only}
Green 2020 {published data only}
Grijalva 2020 {published data only}
-
- Grijalva CG, Zhu Y, Halasa NB, Kim A, Rolfes MA, Steffens A, et al. 432. High concordance between self-collected nasal swabs and saliva samples for detection of SARS-CoV-2. Open Forum Infectious Diseases 2020;7(Supplement 1):S283.
Guo 2020 {published data only}
Guthrie 2020 {published data only}
Guthrie 2021 {published data only}
Hagbom 2021 {published data only}
Hamed 2020 {published data only}
Han 2020 {published data only}
He 2020 {published data only}
-
- He J, Sun Y, Wang L, Wu J, Gong L, Hou S, et al. The virological and epidemiological features of COVID-19 in Anhui, China. Social Science Research Network 2020;NA:no pagination. [DOI: 10.2139/ssrn.3551351] - DOI
Hernandez 2021 {published data only}
-
- Hernandez MM, Banu R, Shrestha P, Patel A, Chen F, Cao LY, et al. Comparison of real-time RT-PCR and RT-PCR/MALDI-TOF methods for SARS-CoV-2 detection in saliva. American Journal of Clinical Pathology 2021;156(SUPPL 1):S9.
Hitzenbichler 2021 {published data only}
Ho 2021 {published data only}
-
- Ho HL, Lin YY, Wang FY, Wu CH, Lee CL, Chou TY. Establishing diagnostic algorithms for SARS-CoV-2 nucleic acid testing in clinical practice. Journal of the Chinese Medical Association 2021;84(12):1120-5. - PubMed
Hoch 2021 {published data only}
-
- Hoch M, Vogel S, Eberle U, Kolberg L, Gruenthaler V, Fingerle V, et al. Feasibility and accuracy of a novel saliva sampling method for large-scale SARS-CoV-2 screening in children < 12 years of age. MedRxiv 2021;NA:no pagination. [DOI: ]
Hofman 2021 {published data only}
Homza 2021 {published data only}
-
- Homza M, Zelena H, Janosek J, Tomaskova H, Jezo E, Kloudova A, et al. Performance of seven SARS-CoV-2 self-tests based on saliva, anterior nasal and nasopharyngeal swabs corrected for infectiousness in real-life conditions: a cross-sectional test accuracy study. Diagnostics (Basel, Switzerland) 2021;11(9):1567. - PMC - PubMed
Hu 2020 {published data only}
Iftner 2022 {published data only}
-
- Iftner T, Iftner A, Pohle D, Martus P. Evaluation of the specificity and accuracy of SARS-CoV-2 rapid antigen self-tests compared to RT-PCR from 1015 asymptomatic volunteers. MedRxiv 2022;NA:no pagination. [DOI: ]
Igloi 2021 {published data only}
Iwasaki 2020 {published data only}
Jamal 2020 {published data only}
Jayaprakasam 2021 {published data only}
-
- Jayaprakasam M, Aggarwal S, Mane A, Saxena V, Rao A, Bandopadhyay B, et al. RNA-extraction-free diagnostic method to detect SARS-CoV-2: an assessment from two states, India. Epidemiology and Infection 2021;149:e245.
Jiang 2020 {published data only}
-
- Jiang G, Ren X, Liu Y, Chen H, Liu W, Guo Z, et al. Application and optimization of RT-PCR in diagnosis of SARS-CoV-2 infection. MedRxiv 2020;NA:1-29.
Kalil 2021 {published data only}
Kandel 2020 {published data only}
Kandel 2021 {published data only}
-
- Kandel CE, Young M, Serbanescu MA, Powis JE, Bulir D, Callahan J, et al. Detection of severe acute respiratory coronavirus virus 2 (SARS-CoV-2) in outpatients: a multicenter comparison of self-collected saline gargle, oral swab, and combined oral-anterior nasal swab to a provider collected nasopharyngeal swab. Infection Control and Hospital Epidemiology 2021;42(11):1340-4. - PMC - PubMed
Kerimov 2021 {published data only}
Kidd 2022 {published data only}
Kim 2020 {published data only}
Kim 2020a {published data only}
Kim 2022 {published data only}
Kinshella 2022 {published data only}
Kitt 2020 {published data only}
-
- Kitt E, Sammons JS, Chiotos K, Coffin SE, Coffin SE, Ballantine A, et al. 425. The utility of paired upper and lower COVID-19 sampling in patients with artificial airways. Open Forum Infectious Diseases 2020;7(Supplement 1):S279.
Kiyasu 2021 {published data only}
Kiyasu 2022 {published data only}
-
- Kiyasu Y, Owaku M, Akashi Y, Takeuchi Y, Narahara K, Mori S, et al. Clinical evaluation of the rapid nucleic acid amplification point-of-care test (Smart Gene SARS-CoV-2) in the analysis of nasopharyngeal and anterior nasal samples. Journal of Infection and Chemotherapy 2022;28(4):543-7. - PMC - PubMed
Kiyasu 2022a {published data only}
-
- Kiyasu Y, Owaku M, Akashi Y, Takeuchi Y, Narahara K, Mori S, et al. Clinical evaluation of the rapid nucleic acid amplification point-of-care test (Smart Gene SARS-CoV-2) in the analysis of nasopharyngeal and anterior nasal samples. Journal of Infection and Chemotherapy 2022;28(4):543-7. - PMC - PubMed
Klein 2021a {published data only}
-
- Klein JAF, Kruger LJ, Tobian F, Gaeddert M, Lainati F, Schnitzler P, et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Medical Microbiology and Immunology 2021;210(4):181-6. - PMC - PubMed
Kobayashi 2021 {published data only}
Kojima 2021 {published data only}
Kritikos 2021 {published data only}
-
- Kritikos A, Caruana G, Brouillet R, Miroz JP, Abed-Maillard S, Stieger G, et al. Sensitivity of rapid antigen testing and RT-PCR performed on nasopharyngeal swabs versus saliva samples in COVID-19 hospitalized patients: results of a prospective comparative trial (RESTART). Microorganisms 2021;9(9):1910. - PMC - PubMed
Kujawski 2020 {published data only}
-
- Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nature Medicine 2020;26(6):861-8. - PubMed
-
- Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, et al. First 12 patients with Coronavirus disease 2019 (COVID-19) in the United States [D]. MedRxiv 2020;NA:1-21. - PubMed
Kwon 2021 {published data only}
-
- Kwon J, Ko E, Cho SY, Lee YH, Jun S, Lee K, et al. Bean extract-based gargle for efficient diagnosing COVID-19 at early-stage using rapid antigen tests: a clinical, prospective, diagnostic study. MedRxiv 2021;NA:no pagination. [DOI: ]
Lai 2020 {published data only}
Lai 2021 {published data only}
Lalli 2021 {published data only}
Landaverde 2022 {published data only}
Lapierre 2021 {published data only}
LeBlanc 2020 {published data only}
LeBlanc 2021 {published data only}
Levican‐Asenjo 2020 {published data only}
-
- Levican-Asenjo JE, Almonacid LI, Valenzuela G, Garcia T, Rojas L, Serrano E, et al. Viral shedding dynamics reveals sputum as a reliable and cost-saving specimen for SARS-CoV-2 diagnosis within the first 10 days since symptom onset: a prospective cohort study. MedRxiv 2020;NA:no pagination. [DOI: 10.1101/2020.08.30.20183889] - DOI
Li 2020 {published data only}
Lin 2020 {published data only}
-
- Lin C, Xiang J, Yan M, Li H, Huang S, Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-CoV-2)-infected pneumonia (COVID-19). Clinical Chemistry and Laboratory Medicine 2020;58(7):1089-94. - PubMed
Lin 2022 {published data only}
-
- Lin J, Frediani JK, Damhorst GL, Sullivan JA, Westbrook A, McLendon K, et al. Where is Omicron? Comparison of SARS-CoV-2 RT-PCR and antigen test sensitivity at commonly sampled anatomic sites over the course of disease. MedRxiv 2022;13(Supplement):S194.
Lindner 2021a {published data only}
Lindner 2021b {published data only}
Lindner 2021c {published data only}
-
- Lindner AK, Nikolai O, Rohardt C, Kausch F, Wintel M, Gertler M, et al. SARS-CoV-2 patient self-testing with an antigen-detecting rapid test: a head-to-head comparison with professional testing. MedRxiv 2021;NA:no pagination. [DOI: ]
Lopez‐Martinez 2020 {published data only}
-
- Lopez-Martinez B, Guzman-Ortiz AL, Nevarez-Ramirez AJ, Parra-Ortega I, Olivar-Lopez VB, Angeles-Floriano T, et al. Saliva as a promising biofluid for SARS-CoV-2 detection during the early stages of infection. Boletin Medico del Hospital Infantil de Mexico 2020;77(5):228-33. - PubMed
Lu 2020 {published data only}
Lui 2020 {published data only}
Lyu 2020 {published data only}
-
- Lyu L, Li L, Liu X, Xie T, Zhou P, Zheng B, et al. Analysis of novel coronavirus nucleic acid detection in different specimens of COVID-19 patients after treatment in Tianjin. Chinese Journal of Microbiology and Immunology 2020;40(6):405-9.
L’Helgouach 2020 {published data only}
-
- L'Helgouach N, Champigneux P, Schneider FS, Molina L, Espeut J, Alali M, et al. Easy COV-Lamp based rapid detection of SARS-CoV-2 in saliva. MedRxiv 2020;NA:no pagination. [DOI: ]
Mahmood 2021 {published data only}
Mak 2022 {published data only}
Malczynski 2020 {published data only}
Manzoor 2020 {published data only}
-
- Manzoor S. Comparison of oropharyngeal and nasopharyngeal swabs for detection of SARS-CoV-2 in patients with COVID-19. Chest 2020;158(4):2473a.
Masia 2021 {published data only}
-
- Masia M, Fernandez-Gonzalez M, Sanchez M, Carvajal M, Garcia JA, Gonzalo-Jimenez N, et al. Nasopharyngeal panbio COVID-19 antigen performed at point-of-care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infectious Diseases 2021;8(3):ofab059. - PMC - PubMed
Masse 2021 {published data only}
Matic 2020 {published data only}
Matic 2020a {published data only}
Michalina 2022 {published data only}
-
- Montaño MA, Bemer MJ, Heller KB, Meisner A, Marfatia Z, Rechkina EA, et al. Performance of anterior nares and tongue swabs for nucleic acid, Nucleocapsid and Spike antigen testing for detecting SARS-CoV-2 against nasopharyngeal PCR and viral culture. International Journal of Infectious Diseases 2022;117:287-94. - PMC - PubMed
Miller 2022 {published data only}
-
- Miller EW, Lamberson CM, Akabari RR, Riddell SW, Middleton FA, Nasr MR, et al. Development and validation of two RT-qpcr diagnostic assays for detecting severe acute respiratory syndrome Coronavirus 2 genomic targets across two specimen types. Journal of Molecular Diagnostics 2022;24(4):294-308. - PMC - PubMed
Miranda‐Ortiz 2021 {published data only}
-
- Miranda-Ortiz H, Fernandez-Figueroa EA, Ruiz-Garcia EB, Munoz-Rivas A, Mendez-Perez A, Mendez-Galvan J, et al. Development of an alternative saliva test for diagnosis of SARS-CoV-2 using TRIzol: adapting to countries with lower incomes looking for a large-scale detection program. PLoS One 2021;16(8):e0255807. - PMC - PubMed
Mittal 2020 {published data only}
Mollaei 2020 {published data only}
Montano 2022 {published data only}
-
- Montano MA, Bemer MJ, Heller KB, Meisner A, Marfatia Z, Rechkina EA, et al. Performance of anterior nares and tongue swabs for nucleic acid, Nucleocapsid, and Spike antigen testing for detecting SARS-CoV-2 against nasopharyngeal PCR and viral culture. International Journal of Infectious Diseases 2022;117:287-94. - PMC - PubMed
Moore 2020a {published data only}
Naito 2021 {published data only}
Nikolai 2021 {published data only}
Oba 2022 {published data only}
Onsongo 2022 {published data only}
Paap 2022 {published data only}
Paliksa 2021 {published data only}
Palmas 2020 {published data only}
-
- Palmas G, Moriondo M, Trapani S, Ricci S, Calistri E, Pisano L, et al. Nasal swab as preferred clinical specimen for COVID-19 testing in children. Pediatric Infectious Disease Journal 2020;39(9):e267-70. - PubMed
Pan 2020 {published data only}
Peng 2020 {published data only}
Perkins 2022 {published data only}
Phan 2022 {published data only}
Pinninti 2020 {published data only}
Qiu 2022 {published data only}
-
- Qiu Y, Lu L, Halven A, Terrio R, Yuldelson S, Dougal N, et al. COVIDFast: a high-throughput and RNA extraction-free method for SARS-CoV-2 detection in swab (SwabFAST) or saliva (SalivaFAST). MedRxiv 2022;NA:no pagination. [DOI: ]
Radbel 2020 {published data only}
Rao 2020a {published data only}
Rao 2020b {published data only}
-
- Rao S, Ambroggio L, Asturias EJ, Bajaj L, Corrado M, Inge T, et al. 408. Evaluation of the negative predictive value of the SARS-CoV-2 PCR respiratory assays in asymptomatic children undergoing surgery. Open Forum Infectious Diseases 2020;7(Supplement 1):S272.
Ren 2021 {published data only}
-
- Ren A, Sohaei D, Zacharioudakis I, Sigal GB, Stengelin M, Mathew A, et al. Ultrasensitive assay for saliva-based SARS-CoV-2 antigen detection. MedRxiv 2021;NA:no pagination. [DOI: ] - PubMed
Ricci 2021 {published data only}
Robinson 2022 {published data only}
Sarinoglu 2021 {published data only}
-
- Sarinoglu RC, Guneser D, Sengel BE, Korten V, Yagci AK. Evaluation of different respiratory samples and saliva for the detection of SARS CoV-2 RNA. Marmara Medical Journal 2021;34(1):51-6.
Sasikala 2021 {published data only}
Savage 2021 {published data only}
Sawano 2021 {published data only}
-
- Sawano M, Takeshita K, Ohno H, Oka H. RT-PCR diagnosis of COVID-19 from exhaled breath condensate: a clinical study. Journal of Breath Research 2021;15(3):037103. - PubMed
Schrom 2022 {published data only}
Schuit 2021 {published data only}
-
- Schuit E, Venekamp RP, Veldhuijzen IK, Van den Bijllaardt W, Pas SD, Stohr J, et al. Accuracy and usability of saliva and nasal rapid antigen self-testing for detection of SARS-CoV-2 infection in the general population: a head-to-head comparison. MedRxiv 2021;NA:no pagination. [DOI: 10.1101/2021.12.08.21267452] - DOI
Singh 2021 {published data only}
-
- Singh K, Arrode-Bruses G, Mansini M, Paden KA, Noto J, Shunevich U, et al. Microbiology - Infectious diseases including COVID-19. Clinical Chemistry and Laboratory Medicine 2021;59(s1):s637-757. - PubMed
Suh 2021 {published data only}
-
- Suh In B, Lim J, Kim Hyo S, Rhim G, Heebum K, Hana K, et al. Development and evaluation of Accupower(r) COVID-19 multiplex real-time RT-PCR kit and Accupower(r) SARS-CoV-2 multiplex real-time RT-PCR kit for SARS-CoV-2 detection in sputum, NPS/OPS, saliva and pooled samples. MedRxiv 2021;NA:no pagination. [DOI: 10.1101/2021.11.21.21264927] - DOI - PMC - PubMed
Sun 2021 {published data only}
Sutjipto 2020 {published data only}
Takeuchi 2021 {published data only}
-
- Takeuchi Y, Akashi Y, Kiyasu Y, Terada N, Kurihara Y, Kato D, et al. Clinical evaluation of a combo rapid antigen test Quicknavi-flu + COVID-19 ag for simultaneous detection of SARS-CoV-2 and influenza viruses. MedRxiv 2021;NA:no pagination. [DOI: ]
Taki 2021 {published data only}
Tallmadge 2021 {published data only}
Tan 2020 {published data only}
Tang 2020 {published data only}
Ter‐Ovanesyan 2021a {published data only}
-
- Ter-Ovanesyan D, Gilboa T, Lazarovits R, Rosenthal A, Yu X, Li JZ, et al. Ultrasensitive measurement of both SARS-CoV-2 RNA and antibodies from saliva. Analytical Chemistry 2021;93(13):5365-70. - PubMed
Ter‐Ovanesyan 2021b {published data only}
-
- Ter-Ovanesyan D, Gilboa T, Lazarovits R, Rosenthal A, Yu Xu G, Li Jonathan Z, et al. Ultrasensitive measurement of both SARS-CoV2 RNA and serology from saliva. MedRxiv 2021;NA:no pagination. [DOI: ] - PubMed
Therchilsen 2020 {published data only}
Thwe 2021 {published data only}
To 2020 {published data only}
Trobajo‐Sanmartin 2021 {published data only}
Truong 2020 {published data only}
Truong 2021 {published data only}
Unsaler 2021 {published data only}
Uribe‐Alvarez 2021 {published data only}
Uwamino 2020 {published data only}
-
- Uwamino Y, Nagata M, Aoki W, Fujimori Y, Nakagawa T, Yokota H, et al. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. Journal of Clinical Pathology 2020;74(1):67-8. - PubMed
Vander 2021 {published data only}
Venekamp 2021 {published data only}
Villota 2021 {published data only}
Vogels 2021 {published data only}
Wang 2020a {published data only}
Wei 2020 {published data only}
Wolfl‐Duchek 2022 {published data only}
Won 2021 {published data only}
Wong 2021 {published data only}
Woodall 2021 {published data only}
-
- Woodall CA, Thornton HV, Anderson EC, Ingle SM, Muir P, Vipond B, et al. Prospective study of the performance of parent-collected nasal and saliva swab samples, compared with nurse-collected swab samples, for the molecular detection of respiratory microorganisms. Microbiology Spectrum 2021;9(3):e0016421. - PMC - PubMed
Wu 2020a {published data only}
Wu 2020b {published data only}
Wyllie 2020 {published data only}
Wyllie 2022 {published data only}
-
- Wyllie AL, Premsrirut PK. Saliva RT-PCR sensitivity over the course of SARS-CoV-2 infection. JAMA 2022;327(2):182-3. - PubMed
Xiao 2020 {published data only}
Xie 2020 {published data only}
Xu 2020 {published data only}
-
- Xu L, Zhang X, Song W, Sun B, Mu J, Dong X, et al. Conjunctival polymerase chain reaction-tests of 2019 novel Coronavirus in patients in Shenyang, China. MedRxiv 2020;NA:no pagination. [DOI: 10.1101/2020.02.23.20024935] - DOI
Ye 2020 (a) {published data only}
-
- Ye G, Li Y, Lu M, Chen S, Luo Y, Wang S, et al. Standardized sampling by the same nurse could improve the positive rate and consistency of the throat swabs to detect COVID-19. Social Science Research Network 2020;NA:no pagination. [DOI: 10.2139/ssrn.3546084] - DOI
Yin 2022 {published data only}
Yokota 2021 {published data only}
Young 2020 {published data only}
Yu 2020 {published data only}
-
- Yu C, Li L, Tuersun Y, Zhao X, Feng Q, Zhang T, et al. Oropharyngeal secretion as alternative for SARS-CoV-2 detection. Journal of Dental Research 2020;99(10):1199-205. - PubMed
Yuan 2020 {published data only}
Zhang 2022 {published data only}
Zheng 2020 {published data only}
Zobrist 2022 {published data only}
Additional references
Alford 2021
-
- Alford J. COVID-19 linked with wider set of symptoms than previously thought – REACT study; 28 September 2021. www.imperial.ac.uk/news/214693/covid-19-linked-with-wider-symptoms-than/ (accessed prior to 1 November 2024).
Arevalo‐Rodriguez 2020
Atieh 2021
Baden 2009
-
- Baden LR, Drazen JM, Kritek PA, Curfman GD, Morrissey S, Campion EW. H1N1 influenza A disease-information for health professionals [H1N1 influenza A disease-information for health professionals]. New England Journal of Medicine 2009;360:2666-2667. [PMID: doi: 10.1056/NEJMe0903992] - PubMed
Bastos 2021
Bhattarai 2018
Blackmore 2020
-
- Blackmore C. The role of population-wide saliva sampling for COVID-19. Rapid response to Covid-19: concerns persist about purpose, ethics, and effect of rapid testing in Liverpool. BMJ (Clinical Research Ed.) 2020;371(41):371. [DOI: 10.1136/bmj.m4690] [URL: www.bmj.com/content/371/bmj.m4690/rr] - DOI - PubMed
Block‐Wheeler 2023
Bossuyt 2015
Brummer 2021
Butler‐Laporte 2021
-
- Butler-Laporte G, Lawandi A, Schiller I, Yao M, Dendukuri N, McDonald EG, et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Internal Medicine 2021;181(3):353-60. [DOI: 10.1001/jamainternmed.2020.8876] - DOI - PMC - PubMed
Centres for Disease Control 2021
-
- Centers for Disease Control. Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html... (accessed 26 February 2021).
Cevik 2021
COVID‐19 Open Access Project (COAP) 2020
-
- COVID-19 Open Access Project (COAP). Living Evidence on COVID-19. ispmbern.github.io/covid-19/living-review/ (accessed 17 February 2020).
Covidence [Computer program]
-
- Covidence. Version (accessed 30 April 2024). Melbourne, Australia: Veritas Health Innovation, 2024. Available at covidence.org.
Crozier 2021
DerSimonian 1986
Dinnes 2022
Ebrahimzadeh 2022
ECDC 2021
-
- European Centre for Disease Prevention and Control. Considerations for the use of saliva as sample material for COVID-19 testing; 3 May 2021. www.ecdc.europa.eu/en/publications-data/considerations-use-saliva-sample... (accessed prior to 2 November, 2024).
ECDC 2022
-
- European Centre for Disease Prevention and Control. Diagnostic testing and screening for SARS-CoV-2. www.ecdc.europa.eu/en/covid-19/surveillance/testing-strategies (accessed 25th January 2023).
Fox 2022
Frazee 2018
-
- Frazee BW, Rodriguez-Hoces de la Guardia A, Alter H, Chen CG, Fuentes EL, Holzer AK, et al. Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Annals of Emergency Medicine 2018;71(4):509-17.e1. [DOI: 10.1016/j.annemergmed.2017.09.010] - DOI - PubMed
Ghafouri‐Fard 2020
Kivela 2021
Kucirka 2020
Lee 2021
Levy 2021
Mallett 2020
-
- Mallett S, Allen AJ, Graziadio S, Taylor SA, Sakai NS, Green K, et al. At what times during infection is SARS-CoV-2 detectable and no longer detectable using RT-PCR-based tests? A systematic review of individual participant data. BMC Medicine 2020;18(1):346. [DOI: 10.1186/s12916-020-01810-8] - DOI - PMC - PubMed
Mane 2022
-
- Mane A, Jain S, Jain A, Nema V, Kurle S, Saxena V, et al. Diagnostic performance of patient self-collected oral swab (tongue and cheek) in comparison with healthcare worker-collected nasopharyngeal swab for severe acute respiratory syndrome coronavirus-2 detection. Acta Pathologica, Microbiologica, et Immunologica Scandinavica 2022;130(11):671-7. [DOI: 10.1111/apm.13266] - DOI - PMC - PubMed
Mayers 2020
-
- Mayers C, Baker K. Impact of false-positives and false-negatives in the UK’s COVID-19 RT-PCR testing programme; 3 June 2020. assets.publishing.service.gov.uk/government/uploads/system/uploads/attac... (accessed prior to 2 November, 2024).
McLaughlin 2022
Miller 2010
Niedrig 2018
ONS 2020
-
- ONS. Coronavirus (COVID-19) Infection Survey pilot: England and Wales, UK: 21 August 2020. ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsan... (accessed 2020).
Plantamura 2021
-
- Plantamura J, Bousquet A, Otto MP, Bigaillon C, Legland AM, Delacour H, et al. Performances, feasibility and acceptability of nasopharyngeal swab, saliva and oral-self sampling swab for the detection of severe acute respiratory syndrome coronavirus 2. European Journal of Clinical Microbiology & Infectious Diseases 2021;40(10):2191-8. [DOI: 10.1007/s10096-021-04269-4] - DOI - PMC - PubMed
Riley 2020
-
- Riley S, Ainslie KEC, Eales O, Walters CE, Wang H, Atchison C, et al. Transient dynamics of SARS-CoV-2 as England exited national lockdown. medRxiv 2020;NA:no pagination. [DOI: 10.1101/2020.08.05.20169078] [URL: https://www.medrxiv.org/content/10.1101/2020.08.05.20169078v1.full.pdf] - DOI
Rucker 2008
Rutjes 2006
Sri 2020
STATA [Computer program]
-
- Stata. Version 17. College Station, TX, USA: StataCorp, 2021. Available at www.stata.com.
Stegeman 2020
Struyf 2022
-
- Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19. Cochrane Database of Systematic Reviews 2022, Issue 5. Art. No: CD013665. [DOI: 10.1002/14651858.CD013665.pub3] - DOI - PMC - PubMed
Tsang 2022
Uwamino 2021
Weiss 2020
Whiting 2011
WHO 2019
-
- World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance; 2 March 2020. iris.who.int/handle/10665/331329 (accessed prior to 2 November, 2024).
WHO 2020a
-
- World Health Organization. Guidelines for the Collection of Clinical Specimens During Field Investigation of Outbreaks; 2000. www.who.int/iris/handle/10665/66348 (accessed 30 March, 2023).
WHO 2020b
-
- World Health Organization. Q&A on infection prevention and control for health care workers caring for patients with suspected or confirmed 2019-nCoV. healthalert.whofreebasics.org/sections/advice-healthcare-workers/qa-on-i... (accessed 2020).
WHO 2021
-
- World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection; 6 October 2021. www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sa... (accessed prior to 2 November 2024).
WHO 2023
-
- World Health Organization. WHO Coronvirus (COVID-19) Dashboard. data.who.int/dashboards/covid19/cases (accessed 16 March 2023).
Wolfel 2020
Yang 2020
Yang 2021
Zimba 2021
References to other published versions of this review
Deeks 2020a
-
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. Art. No: CD013596. [DOI: 10.1002/14651858.CD013596] - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

