The Landscape of ctDNA in Appendiceal Adenocarcinoma
- PMID: 39679931
- PMCID: PMC11790361
- DOI: 10.1158/1078-0432.CCR-24-2474
The Landscape of ctDNA in Appendiceal Adenocarcinoma
Abstract
Purpose: Appendiceal adenocarcinoma is a rare malignancy with distinct histopathologic subtypes and a natural history with metastasis primarily limited to the peritoneum. Little is known about the molecular pathogenesis of appendiceal adenocarcinoma relative to common tumors.
Experimental design: We analyzed molecular data for patients within the Guardant Health database with appendix cancer (n = 718). We then identified patients with appendiceal adenocarcinoma at our institution (from October 2004-September 2022) for whom ctDNA mutation profiling (liquid biopsy) was performed (n = 168) and extracted clinicopathologic and outcomes data. Of these 168 patients, 57 also had tissue-based tumor mutational profiling, allowing for evaluation of concordance between liquid and tissue assays.
Results: The mutational landscape of ctDNA in appendiceal adenocarcinoma is distinct from tissue-based sequencing, with TP53 being the most frequently mutated (46%). Relative to other tumors, appendiceal adenocarcinoma seems less likely to shed ctDNA, with only 38% of patients with metastatic appendiceal adenocarcinoma having detectable ctDNA (OR = 0.26; P < 0.0001 relative to colorectal cancer). When detectable, the median variant allele frequency was significantly lower in appendiceal adenocarcinoma (0.4% vs. 1.3% for colorectal cancer; P ≤ 0.001). High-grade, signet ring, or colonic-type histology, metastatic spread beyond the peritoneum, and TP53 mutation were associated with detectable ctDNA. With respect to clinical translation, patients with detectable ctDNA had worse overall survival (HR = 2.32; P = 0.048). In the Guardant Health cohort, actionable mutations were found in 93 patients (13.0%).
Conclusions: Although metastatic appendiceal adenocarcinoma tumors are less likely to shed tumor DNA into the blood relative to colorectal cancer, ctDNA profiling in appendiceal adenocarcinoma has clinical utility.
©2024 American Association for Cancer Research.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
A.D. reports research support from Novartis, Eisai, Ipsen, Hutchison Pharma, Guardant Health, and Natera; and personal/consulting fees from AbbVie, Crinetics, Hutchison Pharma, Ipsen, Novartis, Personalis, and Voluntis.
B.K. reports Medtronic stock outside the submitted work.
M.O. reports personal fees from Jansen, Bristol-Myers Squibb, Merck, Abbvie, Medimmune, and Takeda and research support from Merck, Bristol-Myers Squibb, Lilly, Nouscom, Medimmune, and Genentech/Roche.
K.R. reports consulting or advisory roles: Abbvie, AstraZeneca, Bayer, Eisai, Daiichi Sankyo, Sanofi, Seagen. Research funding (institutional): Abbvie, Bayer, Roche/Genentech, Guardant Health, Daiichi Sankyo/Astra Zeneca, Janssen, HiberCell, Innovent, Merck Serono, Seagen, Xencor.
A.U. reports consulting with Bayer pharmaceutical.
L.M.D. is an employee of Guardant Health and holds stock options as such.
J.P.S. reports consulting with Engine Biosciences and NaDeNo Nanoscience.
Figures



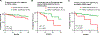
Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer.Gastroenterology. 2019 Jan;156(1):108-118.e4. doi: 10.1053/j.gastro.2018.09.022. Epub 2018 Sep 19. Gastroenterology. 2019. PMID: 30240661 Free PMC article.
-
Prognostic Significance of Circulating Tumor DNA Mutations in Gastrointestinal Stromal Tumors: A Systematic Review and Meta-analysis Based on Time-To-Event Data.J Gastrointest Cancer. 2025 Jul 15;56(1):153. doi: 10.1007/s12029-025-01271-3. J Gastrointest Cancer. 2025. PMID: 40665034
-
Elevated ctDNA Tumor Fraction Is Associated with Improved Mutation Detection but Worse Overall Survival in Advanced Non-Small Cell Lung Cancer: A Lung-MAP Study.Clin Cancer Res. 2025 Aug 14;31(16):3550-3561. doi: 10.1158/1078-0432.CCR-24-3658. Clin Cancer Res. 2025. PMID: 40465842
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
Cited by
-
Applications and advances of multi-omics technologies in gastrointestinal tumors.Front Med (Lausanne). 2025 Jul 23;12:1630788. doi: 10.3389/fmed.2025.1630788. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40771479 Free PMC article. Review.
References
-
- Fernandez RN, Daly JM, Pseudomyxoma peritonei. Archives of surgery 115, 409–414 (1980). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

