CFI-1 functions unilaterally to restrict gap junction formation in C. elegans
- PMID: 39679967
- PMCID: PMC11829774
- DOI: 10.1242/dev.202955
CFI-1 functions unilaterally to restrict gap junction formation in C. elegans
Abstract
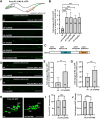
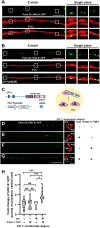
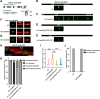
Electrical coupling is vital to neural communication, facilitating synchronized activity among neurons. Despite its significance, the precise mechanisms governing the establishment of gap junction connections between specific neurons remain elusive. Here, we identified that the PVC interneuron in Caenorhabditis elegans forms gap junction connections with the PVR interneuron. The transcriptional regulator CFI-1 (ARID3) is specifically expressed in the PVC but not PVR interneuron. Reducing cfi-1 expression in the PVC interneuron leads to enhanced gap junction formation in the PVR neuron, while ectopic expression of cfi-1 in the PVR neuron restores the proper level of gap junction connections in the PVC neuron, along with the normal touch response. These findings unveil the pivotal role of CFI-1 in bidirectionally regulating the formation of gap junctions within a specific neuronal pair, shedding light on the intricate molecular mechanisms governing neuronal connectivity in vivo.
Keywords: C. elegans; CFI-1/ARID3; Gap junction; Neural circuitry; Transcription factor.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

